9GT4
 
 | |
9GT7
 
 | |
1MMV
 
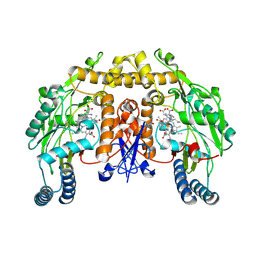 | | Rat neuronal NOS heme domain with NG-propyl-L-arginine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-PROPYL-L-ARGININE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
2P3J
 
 | |
9GKO
 
 | | Structure of 6mer pore intermediate of Sticholysin II (StnII) toxin in lipid nanodiscs | | Descriptor: | DELTA-stichotoxin-She4b, sphingomyelin | | Authors: | Martin Benito, J, Santiago, C, Carlero, D, Arranz, R. | | Deposit date: | 2024-08-25 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Elucidating the structure and assembly mechanism of actinoporin pores in complex membrane environments.
Sci Adv, 11, 2025
|
|
1MG3
 
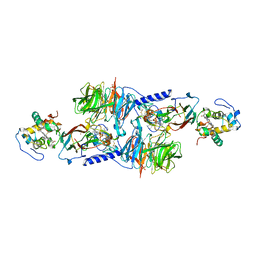 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
9G49
 
 | |
7S1U
 
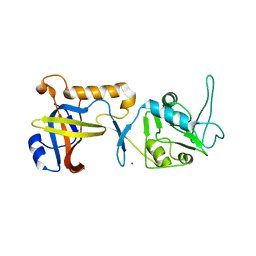 | | Structure of human POT1C | | Descriptor: | Protection of telomeres protein 1, ZINC ION | | Authors: | Aramburu, T, Skordalakes, E. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | POT1-TPP1 binding stabilizes POT1, promoting efficient telomere maintenance.
Comput Struct Biotechnol J, 20, 2022
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
9GOU
 
 | | Cryo-EM structure of acylaminoacyl-peptidase in complex with dichlorvos | | Descriptor: | Acylamino-acid-releasing enzyme, dimethyl hydrogen phosphite | | Authors: | Kiss-Szeman, A.J, Traore, D, Jakli, I, Harmat, V, Menyhard, D.K, Perczel, A. | | Deposit date: | 2024-09-06 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Ligand binding Pro-miscuity of acylpeptide hydrolase, structural analysis of a detoxifying serine hydrolase.
Protein Sci., 34, 2025
|
|
1MID
 
 | |
9GNE
 
 | | CryoEM structure of mammalian AAP in complex with acetyl-alanyl-chloromethylketone | | Descriptor: | Acylamino-acid-releasing enzyme, ~{N}-[(2~{S},3~{S})-3-oxidanylbutan-2-yl]ethanamide | | Authors: | Kiss-Szeman, A.J, Jakli, I, Hosogi, N, Banoczi, Z, Harmat, V, Memyhard, D.K, Perczel, A. | | Deposit date: | 2024-09-02 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand binding Pro-miscuity of acylpeptide hydrolase, structural analysis of a detoxifying serine hydrolase.
Protein Sci., 34, 2025
|
|
5FK8
 
 | |
9G4D
 
 | |
7SEN
 
 | |
7SSM
 
 | | Crystal structure of human STING R232 in complex with compound 11 | | Descriptor: | 2-({[(8R)-pyrazolo[1,5-a]pyrimidine-3-carbonyl]amino}methyl)-1-benzofuran-7-carboxylic acid, Stimulator of interferon genes protein | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Non-Nucleotide Small-Molecule STING Agonists via Chemotype Hybridization.
J.Med.Chem., 65, 2022
|
|
1MNV
 
 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
9H83
 
 | | Small circular RNA dimer - Class 2 | | Descriptor: | Circular RNA, Complementary strand | | Authors: | McRae, E.K, Kristoffersen, E.L, Holliger, P, Andersen, E.S. | | Deposit date: | 2024-10-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Roles of dimeric intermediates in RNA-catalyzed rolling circle synthesis.
Nucleic Acids Res., 53, 2025
|
|
9H8A
 
 | | Small circular RNA dimer - Class 5 | | Descriptor: | Circular RNA, Complementary strand | | Authors: | McRae, E.K, Kristoffersen, E.L, Holliger, P, Andersen, E.S. | | Deposit date: | 2024-10-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Roles of dimeric intermediates in RNA-catalyzed rolling circle synthesis.
Nucleic Acids Res., 53, 2025
|
|
1MO6
 
 | |
1MIJ
 
 | |
9GWZ
 
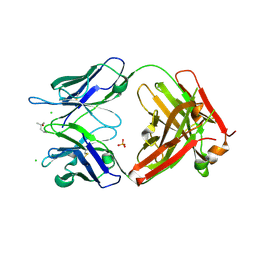 | | Crystal structure of 23ME-00610 Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23ME-00610 Fab (heavy), 23ME-00610 Fab (light), ... | | Authors: | Huang, Y.M, Ganichkin, O.M. | | Deposit date: | 2024-09-27 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Mabs, 16, 2024
|
|
1MIT
 
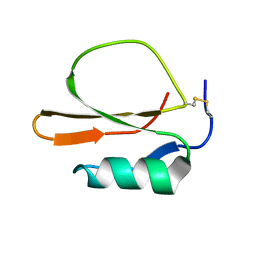 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
9HVP
 
 | | Design, activity and 2.8 Angstroms crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease | | Descriptor: | HIV-1 Protease, benzyl [(1R,4S,6S,9R)-4,6-dibenzyl-5-hydroxy-1,9-bis(1-methylethyl)-2,8,11-trioxo-13-phenyl-12-oxa-3,7,10-triazatridec-1-yl]carbamate | | Authors: | Neidhart, D.J, Erickson, J. | | Deposit date: | 1990-11-06 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease.
Science, 249, 1990
|
|
1LZT
 
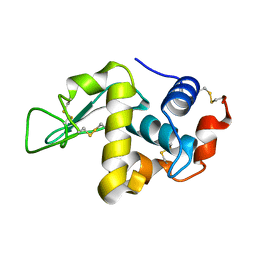 | | REFINEMENT OF TRICLINIC LYSOZYME | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Hodsdon, J.M, Brown, G.M, Sieker, L.C, Jensen, L.H. | | Deposit date: | 1985-04-01 | | Release date: | 1985-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Refinement of triclinic lysozyme: I. Fourier and least-squares methods.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
