7ZG2
 
 | | BRD4 in complex with Acetyl-Lys | | Descriptor: | (2S)-2,6-diacetamido-N-methyl-hexanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFT
 
 | | BRD4 in complex with FragLite33 | | Descriptor: | 3-azanyl-5-bromanyl-1-methyl-pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG1
 
 | | BRD4 in complex with PepLite-Tyr | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, Nalpha-acetyl-N-(3-bromoprop-2-yn-1-yl)-L-tyrosinamide | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7Z9Y
 
 | | BRD4 in complex with FragLite2 | | Descriptor: | 4-IODOPYRAZOLE, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZE7
 
 | | BRD4 in complex with FragLite21 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7Z9W
 
 | | BRD4 in complex with FragLite1 | | Descriptor: | 4-bromo-1H-pyrazole, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.988 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZA8
 
 | | BRD4 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
4JM5
 
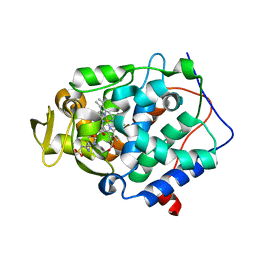 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2-Amino-5-methylthiazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
6A73
 
 | | Complex structure of CSN2 with IP6 | | Descriptor: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | Authors: | Liu, L, Li, D, Rao, F, Wang, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4JM6
 
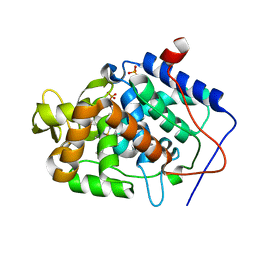 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,4-diaminopyrimidine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
8XSU
 
 | |
5VZU
 
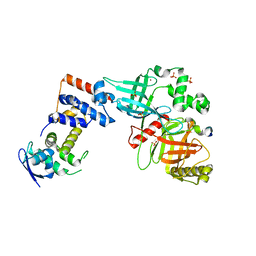 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BZH
 
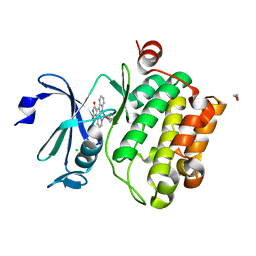 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1, ... | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Ruthenium Organometallic Ligands
To be Published
|
|
1DWS
 
 | | PHOTOLYZED CARBONMONOXY MYOGLOBIN (HORSE HEART) | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1DWR
 
 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
9IWF
 
 | |
5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
4JM8
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,6-diaminopyridine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JPL
 
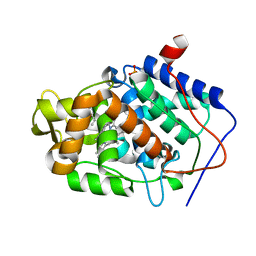 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Azaindole | | Descriptor: | 1H-pyrrolo[3,2-b]pyridine, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQK
 
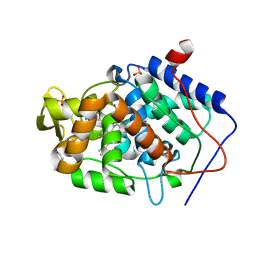 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2-(2-aminopyridin-1-ium-1-yl)ethanol | | Descriptor: | 2-(2-aminopyridin-1-ium-1-yl)ethanol, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JPT
 
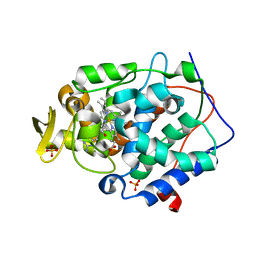 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with quinazoline-2,4-diamine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
2HB4
 
 | | Structure of HIV Protease NL4-3 in an Unliganded State | | Descriptor: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | Authors: | Heaslet, H, Tam, K, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3RS1
 
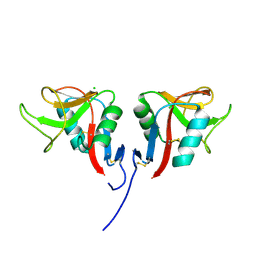 | | Mouse C-type lectin-related protein Clrg | | Descriptor: | C-type lectin domain family 2 member I, CHLORIDE ION | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
4IB7
 
 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyltrimethylammonium (DTAC) | | Descriptor: | DODECANE-TRIMETHYLAMINE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
