8OPL
 
 | |
5TJ2
 
 | |
8OPB
 
 | |
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
8OJ2
 
 | |
5T8Q
 
 | |
8OJ1
 
 | |
8BXU
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
5TJ3
 
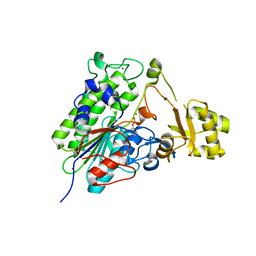 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
8OPA
 
 | |
8OPE
 
 | |
8OPF
 
 | |
8OPK
 
 | |
8A96
 
 | | SARS Cov2 Spike RBD in complex with Fab47 | | Descriptor: | Fab47 Heavy chain (variable domain), Fab47 Light chain (variable domain), Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A99
 
 | | SARS Cov2 Spike in 1-up conformation complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
5NGQ
 
 | | Bicyclic antimicrobial peptides | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, DLS-PRO-ALD-CYS-TYD-ALA-CYD-LYS-ALA, ... | | Authors: | Di Bonaventura, I, Jin, X, Visini, R, Michaud, G, Robadey, M, Koehler, T, van Delden, C, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Chemical space guided discovery of antimicrobial bridged bicyclic peptides against Pseudomonas aeruginosa and its biofilms.
Chem Sci, 8, 2017
|
|
8A94
 
 | | SARS CoV2 Spike in the 2-up state in complex with Fab47. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain Variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
6S5S
 
 | | Cfucosylated second generation peptide dendrimer SBD8 bound to Fucose binding Lectin LecB (PA-IIL) from Pseudomonas aeruginosa at 1.43 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | X-Ray Crystal Structure of a Second Generation Peptide Dendrimer in Complex with Pseudomonas aeruginosa Lectin LecB
Helv.Chim.Acta, 2019
|
|
8A95
 
 | | SARS Cov2 Spike RBD in complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
6SNI
 
 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
6U2H
 
 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2LMO
 
 | |
2LMQ
 
 | |
2LMN
 
 | |
2LMP
 
 | |
