8KA3
 
 | |
8KA5
 
 | |
1ZWX
 
 | | Crystal Structure of SmcL | | Descriptor: | GLYCEROL, PHOSPHATE ION, sphingomyelinase-c | | Authors: | Openshaw, A.E.A, Race, P.R, Monzo, H.J, Vasquez-Boland, J.A, Banfield, M.J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria.
J.Biol.Chem., 280, 2005
|
|
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7SUV
 
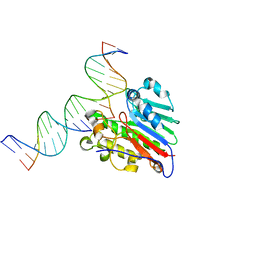 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7TC2
 
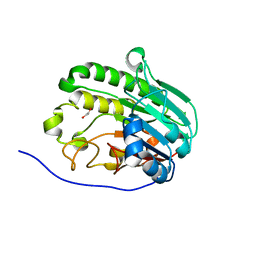 | | Human APE1 in complex with 5-nitroindole-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7TC3
 
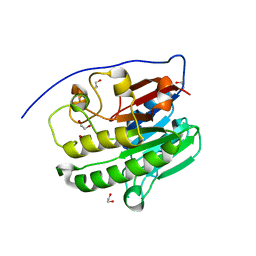 | | Human APE1 in the apo form | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7TR7
 
 | | APE1 product complex with abasic ssDNA | | Descriptor: | DNA (5'-D(P*(3DR)P*CP*GP*AP*TP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2022-01-28 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into AP-endonuclease 1 cleavage of abasic sites at stalled replication fork mimics.
Nucleic Acids Res., 51, 2023
|
|
3G8V
 
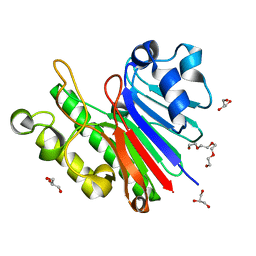 | |
3G91
 
 | | 1.2 Angstrom structure of the exonuclease III homologue Mth0212 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-12 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
2DDT
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with magnesium ion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ago, H, Oda, M, Tsuge, H, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DNJ
 
 | |
2DDS
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with cobalt ion | | Descriptor: | COBALT (II) ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
3I41
 
 | |
3I48
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound magnesium ions | | Descriptor: | Beta-hemolysin, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
to be published, 2009
|
|
3I5V
 
 | |
3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3I46
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound calcium ions | | Descriptor: | Beta-hemolysin, CALCIUM ION, CHLORIDE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
To be Published
|
|
2DDR
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with calcium ion | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2F1N
 
 | |
4FPV
 
 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FVA
 
 | | Crystal structure of truncated Caenorhabditis elegans TDP2 | | Descriptor: | 1,2-ETHANEDIOL, 5'-tyrosyl-DNA phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GZ0
 
 | |
1AKO
 
 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
4GZ2
 
 | | Mus Musculus Tdp2 excluded ssDNA complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*AP*AP*TP*TP*CP*G)-3'), FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
