1PX4
 
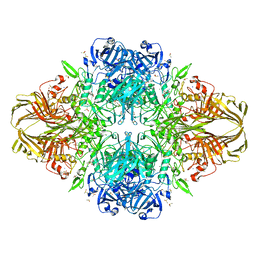 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) WITH IPTG BOUND | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
7CWD
 
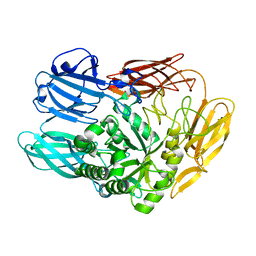 | |
7PR6
 
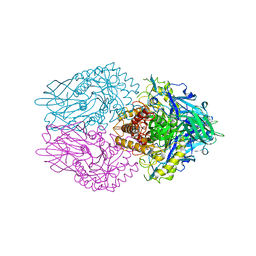 | | Crystal structure of E. coli beta-glucuronidase in complex with covalent inhibitor ME727 | | Descriptor: | (2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4V41
 
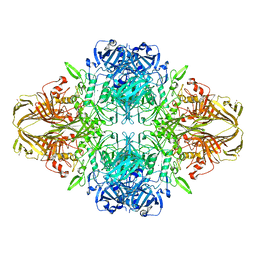 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
6X1Q
 
 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
4V45
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
4V40
 
 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
6XXW
 
 | | Structure of beta-D-Glucuronidase for Dictyoglomus thermophilum. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucuronidase, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Thioglycoligation of aromatic thiols using a natural glucuronide donor.
Org.Biomol.Chem., 18, 2020
|
|
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
7SF2
 
 | | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-02 | | Release date: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus
To Be Published
|
|
1F4H
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
8BK7
 
 | |
8BKG
 
 | |
8BK8
 
 | |
1HN1
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION, SODIUM ION | | Authors: | Juers, D.H, Matthews, B.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions.
J.Mol.Biol., 311, 2001
|
|
5C70
 
 | | The structure of Aspergillus oryzae beta-glucuronidase | | Descriptor: | Glucuronidase | | Authors: | Sun, H.L, Lv, B, Huang, S, Sun, Q.F, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enhancing the Thermostability of beta-Glucuronidase by Rationally Redesigning the Catalytic Domain Based on Sequence Alignment Strategy
Ind Eng Chem Res, 55, 2016
|
|
8Q7Y
 
 | |
5C71
 
 | | The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Glucuronidase, alpha-D-glucopyranuronic acid | | Authors: | Sun, H.L, Lv, B, Huang, S, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-guided engineering of the substrate specificity of a fungal beta-glucuronidase toward triterpenoid saponins.
J.Biol.Chem., 293, 2018
|
|
5A1A
 
 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
8Q51
 
 | | Beta-galactosidase from Bacillus circulans conformational state 2 | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-07 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
8Q2H
 
 | | beta-galactosidase from Bacillus circulans | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
8Q4Y
 
 | | Beta-galactosidase from Bacillus circulans conformational state 1 | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-07 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
4JKK
 
 | |
5Z19
 
 | |
