2OLP
 
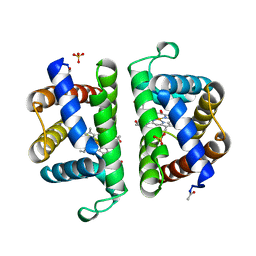 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2007-01-19 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
2DMV
 
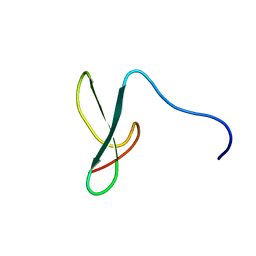 | | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch) | | Descriptor: | Itchy homolog E3 ubiquitin protein ligase | | Authors: | Ohnishi, S, Paakkonen, K, Tochio, N, Tomizawa, T, Koshiba, S, Inoue, M, Guntert, P, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch)
To be Published
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
2ACL
 
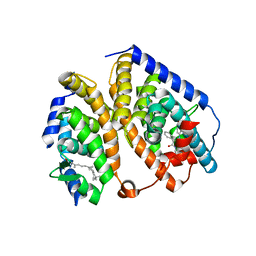 | | Liver X-Receptor alpha Ligand Binding Domain with SB313987 | | Descriptor: | 1-BENZYL-3-(4-METHOXYPHENYLAMINO)-4-PHENYLPYRROLE-2,5-DIONE, Oxysterols receptor LXR-alpha, RETINOIC ACID, ... | | Authors: | Jaye, M.C, Krawiec, J.A, Campobasso, N, Smallwood, A, Qiu, C, Lu, Q, Kerrigan, J.J. | | Deposit date: | 2005-07-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of substituted maleimides as liver x receptor agonists and determination of a ligand-bound crystal structure.
J.Med.Chem., 48, 2005
|
|
2YRN
 
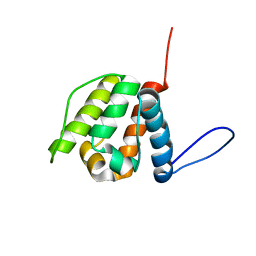 | | Solution structure of the CH domain from Human Neuron navigator 2 | | Descriptor: | Neuron navigator 2 isoform 4 | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Nakamura, Y, Furukawa, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from Human Neuron navigator 2
To be Published
|
|
8QQL
 
 | |
5BTQ
 
 | | Crystal structure of human heme oxygenase 1 H25R with biliverdin bound | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1, SULFATE ION | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
5AIU
 
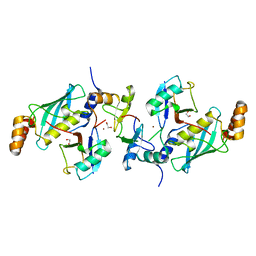 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
8A0W
 
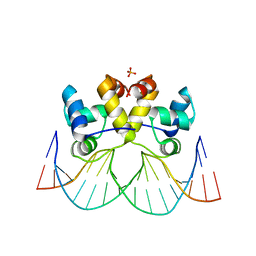 | |
3VGO
 
 | |
1QNU
 
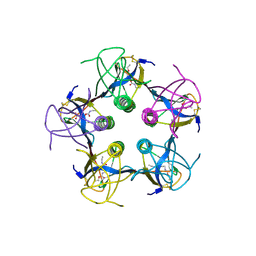 | | Shiga-Like Toxin I B Subunit Complexed with the Bridged-Starfish Inhibitor | | Descriptor: | ETHYL-CARBAMIC ACID METHYL ESTER, METHYL-CARBAMIC ACID ETHYL ESTER, Shiga toxin 1 variant B subunit, ... | | Authors: | Pannu, N.S, Hayakawa, K, Read, R.J. | | Deposit date: | 1999-10-21 | | Release date: | 2000-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands.
Nature, 403, 2000
|
|
6LTY
 
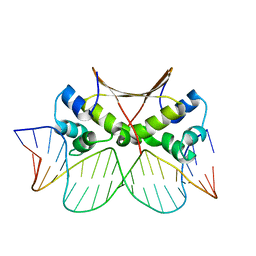 | | DNA bound antitoxin HigA3 | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*GP*AP*GP*AP*TP*AP*TP*AP*AP*CP*CP*TP*AP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*TP*AP*GP*GP*TP*TP*AP*TP*AP*TP*CP*TP*CP*GP*TP*GP*G)-3'), Putative antitoxin HigA3 | | Authors: | Park, J.Y, Lee, B.J. | | Deposit date: | 2020-01-23 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Induced DNA bending by unique dimerization of HigA antitoxin.
Iucrj, 7, 2020
|
|
6LTZ
 
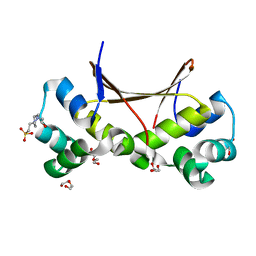 | |
4J9D
 
 | |
4J9B
 
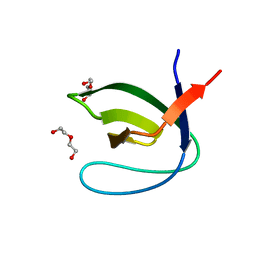 | |
4J9F
 
 | |
4J9E
 
 | |
5AIT
 
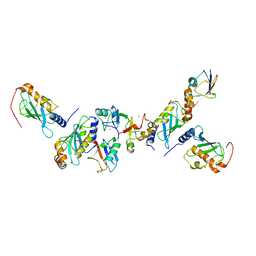 | | A complex of of RNF4-RING domain, UbeV2, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 N, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
8CRP
 
 | |
8CRN
 
 | |
4JJD
 
 | |
4JJB
 
 | |
4JJC
 
 | |
3EG2
 
 | | Crystal structure of the N114Q mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EGU
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1, SULFATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
