1K1E
 
 | |
1K25
 
 | |
1P71
 
 | | Anabaena HU-DNA corcrystal structure (TR3) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', DNA-binding protein HU | | Authors: | Swinger, K.S, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1K1S
 
 | | Crystal Structure of DinB from Sulfolobus solfataricus | | Descriptor: | DBH protein | | Authors: | Silvian, L.F, Toth, E.A, Pham, P, Goodman, M.F, Ellenberger, T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus.
Nat.Struct.Biol., 8, 2001
|
|
1PA2
 
 | | ARABIDOPSIS THALIANA PEROXIDASE A2 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PEROXIDASE, ... | | Authors: | Henriksen, A. | | Deposit date: | 1999-05-18 | | Release date: | 2000-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification.
Plant Mol.Biol., 44, 2000
|
|
1P3M
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
2WQG
 
 | | SAP domain from Tho1: L31W (fluorophore) mutant | | Descriptor: | PROTEIN THO1 | | Authors: | Dodson, C.A, Ferguson, N, Rutherford, T.J, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2009-08-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Engineering a Two-Helix Bundle Protein for Folding Studies.
Protein Eng.Des.Sel., 23, 2010
|
|
4DHJ
 
 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
1JOG
 
 | | Structure of HI0074 from Heamophilus Influenzae reveals the fold of a substrate binding domain of a nucleotidyltransferase | | Descriptor: | HYPOTHETICAL PROTEIN HI0074 | | Authors: | Lehmann, C, Lim, K, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-29 | | Release date: | 2002-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The HI0073/HI0074 protein pair from Haemophilus influenzae is a member of a new nucleotidyltransferase family: Structure, sequence analyses, and
solution studies
Proteins, 50, 2003
|
|
2WHR
 
 | | Crystal structure of acetylcholinesterase in complex with K027 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.-G, Schneider, G, Pang, Y.-P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
1JRK
 
 | | Crystal Structure of a Nudix Protein from Pyrobaculum aerophilum Reveals a Dimer with Intertwined Beta Sheets | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nudix homolog | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8A4M
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 8 (JMV-7061) | | Descriptor: | (2~{S})-2-[bis(1~{H}-imidazol-4-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Sannio, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
2JWQ
 
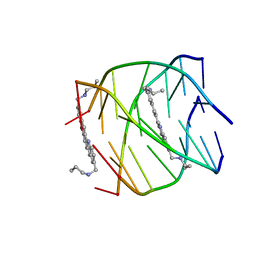 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
8QQH
 
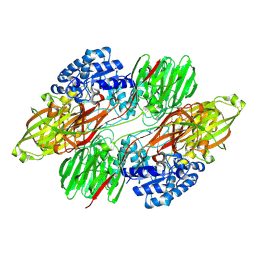 | |
4DTM
 
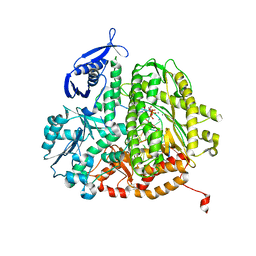 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddG/dC as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DU4
 
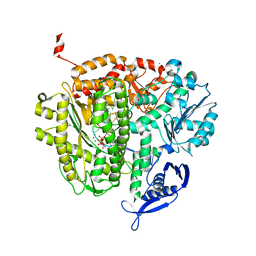 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite dT with 3-Deaza-adenine at the N-3 Position of Primer Strand | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DV4
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DR2
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
2WJM
 
 | | Lipidic sponge phase crystal structure of the photosynthetic reaction centre from Blastochloris viridis (low dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Woehri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
4EDT
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
4EIG
 
 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4EIZ
 
 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
7ZW4
 
 | | Crystal structure of Talin R7R8 domains with Caskin-2 LD-peptide | | Descriptor: | Caskin-2, Talin-1 | | Authors: | Celie, P.H.N, Joosten, R.P, Sonnenberg, A, Perrakis, A. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Caskin2 is a novel talin- and Abi1-binding protein that promotes cell motility.
J.Cell.Sci., 137, 2024
|
|
4EDR
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
