3PBA
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with monosulfate tetrabromo-bisphenol A (MonoTBBPA) | | Descriptor: | 2,6-dibromo-4-[2-(3,5-dibromo-4-hydroxyphenyl)propan-2-yl]phenyl hydrogen sulfate, Peroxisome proliferator-activated receptor gamma, S-1,2-PROPANEDIOL | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2010-10-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Novel Ligands of ER{alpha}, Er{beta}, and PPAR{gamma}: The Case of Halogenated Bisphenol A and Their Conjugated Metabolites.
Toxicol Sci, 122, 2011
|
|
1XRI
 
 | | X-ray structure of a putative phosphoprotein phosphatase from Arabidopsis thaliana gene AT1G05000 | | Descriptor: | At1g05000, SULFATE ION | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-10-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000.
Proteins, 73, 2008
|
|
3P0I
 
 | |
2ONF
 
 | |
3P1F
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand
To be Published
|
|
2OQ2
 
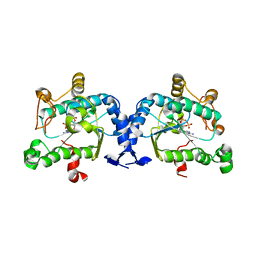 | | Crystal structure of yeast PAPS reductase with PAP, a product complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phosphoadenosine phosphosulfate reductase | | Authors: | Yu, Z, Fisher, A.J. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 3'-phosphoadenosine-5'-phosphosulfate reductase complexed with adenosine 3',5'-bisphosphate.
Biochemistry, 47, 2008
|
|
2OQM
 
 | |
3P3A
 
 | |
2ORD
 
 | |
2OTM
 
 | |
2OU5
 
 | |
2OUW
 
 | |
3P6G
 
 | |
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQS
 
 | | Crystal structure of importin-alpha bound to a CLIC4 NLS peptide | | Descriptor: | Importin subunit alpha-2, peptide of Chloride intracellular channel protein 4 | | Authors: | Mynott, A.V, Brown, L.J, Harrop, S.J, Curmi, P.M.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of importin-alpha bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4
Febs J., 278, 2011
|
|
3P89
 
 | | FXR bound to a quinolinecarboxylic acid | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)quinoline-2-carboxylic acid, Farnesoid X receptor, Nuclear receptor coactivator 1, ... | | Authors: | Madauss, K.P, Williams, S.P, Deaton, D.N. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ORU
 
 | |
3OSD
 
 | |
3P92
 
 | |
2OZH
 
 | |
2P2S
 
 | |
3P1H
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23K/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, thermonuclease | | Authors: | Clark, I.A, Caro, J.A, Sue, G, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-09-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pressure unfolding effects of artificial cavities in proteins.
To be Published
|
|
3OP7
 
 | |
2AE5
 
 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase, SULFATE ION | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2OOK
 
 | |
