5S9X
 
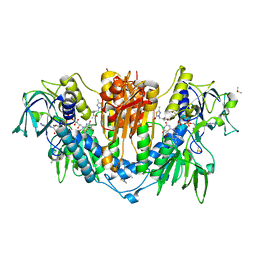 | | PanDDA analysis group deposition -- Crystal Structure of Trypanosoma brucei Trypanothione reductase in complex with Z1899842917 | | Descriptor: | 3-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]-5-methyl-1,3,4-thiadiazol-2(3H)-one, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fiorillo, A, Ilari, A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Innovative Approach for a Classic Target: Fragment Screening on Trypanothione Reductase Reveals New Opportunities for Drug Design.
Front Mol Biosci, 9, 2022
|
|
5S9W
 
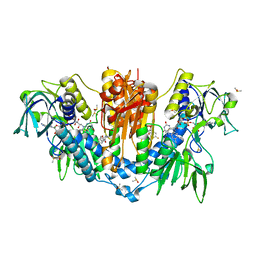 | |
5SA1
 
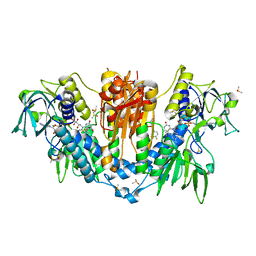 | |
7MK0
 
 | |
5FXT
 
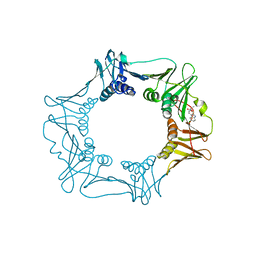 | |
1QKY
 
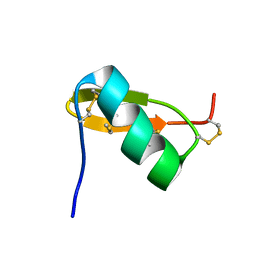 | | Solution structure of PI7, a non toxic peptide isolated from the scorpion Pandinus Imperator. | | Descriptor: | TOXIN 7 FROM PANDINUS IMPERATOR | | Authors: | Delepierre, M, Prochnicka-Chalufour, A, Boisbouvier, J, Possani, L.D. | | Deposit date: | 1999-08-17 | | Release date: | 2000-02-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Pi7, an orphan peptide from the scorpion Pandinus imperator: a 1H-NMR analysis using a nano-NMR Probe.
Biochemistry, 38, 1999
|
|
7MF4
 
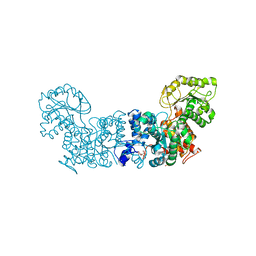 | |
1SQ5
 
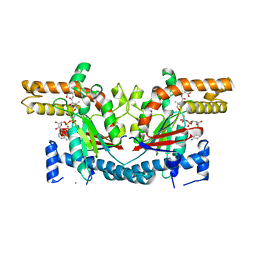 | | Crystal Structure of E. coli Pantothenate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Pantothenate kinase | | Authors: | Ivey, R.A, Zhang, Y.-M, Virga, K.G, Hevener, K, Lee, R.E, Rock, C.O, Jackowski, S, Park, H.-W. | | Deposit date: | 2004-03-17 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the pantothenate kinase.ADP.pantothenate ternary complex reveals the relationship between the binding sites for substrate, allosteric regulator, and antimetabolites.
J.Biol.Chem., 279, 2004
|
|
1OD6
 
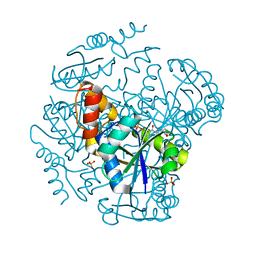 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N2O
 
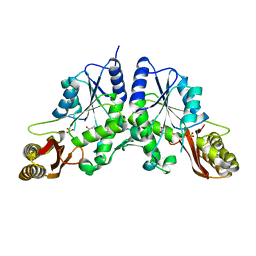 | |
5QOO
 
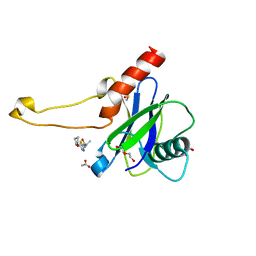 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000144a | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-{5-[(2S)-oxolan-2-yl]-1,3,4-thiadiazol-2(3H)-ylidene}propanamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HLQ
 
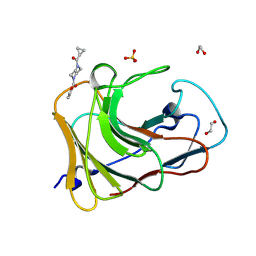 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z32327641 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HO7
 
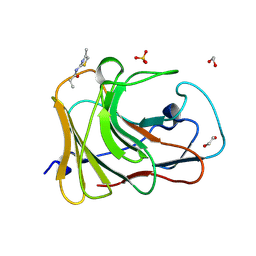 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z30820160 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-(4-methyl-1,3-thiazol-2-yl)propanamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HLM
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z29077827 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HNC
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z220996120 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-[(1S)-1-(pyridin-3-yl)ethyl]cyclopropanecarboxamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HMY
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z111782404 | | Descriptor: | (2S)-2-[([1,1'-biphenyl]-4-yl)oxy]propanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMJ
 
 | | Trypanothione reductase | | Descriptor: | BROMIDE ION, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fiorillo, A, Ilari, A. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Innovative Approach for a Classic Target: Fragment Screening on Trypanothione Reductase Reveals New Opportunities for Drug Design.
Front Mol Biosci, 9, 2022
|
|
1TFU
 
 | |
5QRS
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z1891773393 | | Descriptor: | CADMIUM ION, N-{4-[(1S)-1-aminoethyl]phenyl}cyclopropanecarboxamide, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QS4
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z30820160 | | Descriptor: | CADMIUM ION, N-(4-methyl-1,3-thiazol-2-yl)propanamide, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QXP
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with TCJ732 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5QXQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with TCJ779 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5QXT
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with JKH47 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5RGT
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4439011607 (Mpro-x2540) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(5-tert-butyl-1,2-oxazol-3-yl)propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001558a | | Descriptor: | 3-[4-(4-hydroxyphenyl)phenyl]propanoic acid, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
