3ZKG
 
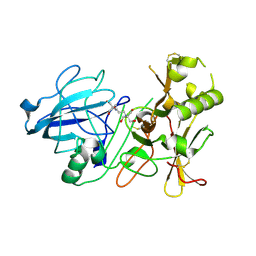 | | BACE2 MUTANT APO STRUCTURE | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZL7
 
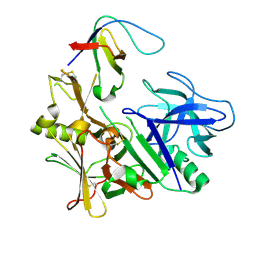 | | BACE2 FYNOMER COMPLEX | | Descriptor: | BETA-SECRETASE 2, FYNOMER 2B-H11 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SFC
 
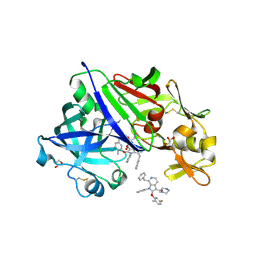 | | Structure-Based Optimization of Potent 4- and 6-Azaindole-3-Carboxamides as Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Renin, ... | | Authors: | Scheiper, B, Matter, H, Steinhagen, H, Bocskei, Z, Fleury, V, McCort, G. | | Deposit date: | 2011-06-13 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of potent 4- and 6-azaindole-3-carboxamides as renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ZKI
 
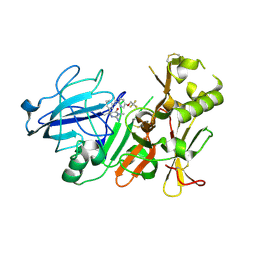 | | BACE2 MUTANT STRUCTURE WITH LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SKG
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-(1,2,3,4-tetrahydro-3-isoquinolinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(3R)-1,2,3,4-tetrahydroisoquinolin-3-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-06-22 | | Release date: | 2011-09-07 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Synthesis and in vivo evaluation of cyclic diaminopropane BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ZKQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZOV
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-02-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZMG
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(4R)-2-azanylidene-5,5-bis(fluoranyl)-4-methyl-1,3-oxazinan-4-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
4AA8
 
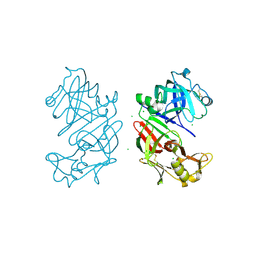 | | Bovine chymosin at 1.8A resolution | | Descriptor: | CHLORIDE ION, CHYMOSIN | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4APR
 
 | |
3S2O
 
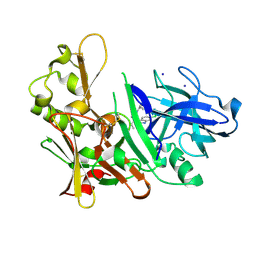 | | Fragment based discovery and optimisation of bace-1 inhibitors | | Descriptor: | (3S)-3-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]pentanamide, Beta-secretase 1, IODIDE ION | | Authors: | Madden, J, Godemann, R, Smith, M.A, Hallett, D, Barker, J, Kraemer, J. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3S7M
 
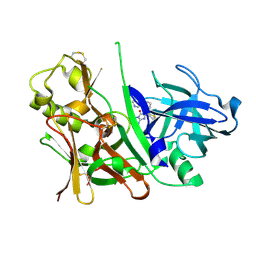 | |
3ZKX
 
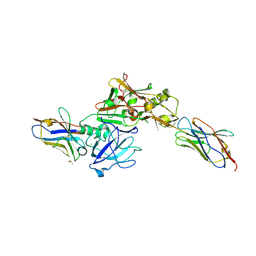 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QBH
 
 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QS1
 
 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
4ACU
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 14 | | Descriptor: | (8S)-3,3-DIFLUORO-8-(2'-FLUORO-3'-METHOXYBIPHENYL-3-YL)-8-PYRIDIN-4-YL-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
3WB5
 
 | | Crystal Structure of beta secetase in complex with (6S)-2-amino-3,6-dimethyl-6-[(1R,2R)-2-phenylcyclopropyl]-3,4,5,6-tetrahydropyrimidin-4-one | | Descriptor: | (6S)-2-amino-3,6-dimethyl-6-[(1R,2R)-2-phenylcyclopropyl]-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Yonezawa, S, Fujiwara, K, Yamamoto, T, Hattori, K, Yamakawa, H, Muto, C, Hosono, M, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors III: Effective investigation of the binding mode by combinational use of X-ray analysis, isothermal titration calorimetry and theoretical calculations
Bioorg.Med.Chem., 21, 2013
|
|
3T7Q
 
 | |
3PI5
 
 | | Crystal Structure of Human Beta Secretase in Complex with BFG356 | | Descriptor: | (3S,4S,5R)-3-(3-bromo-4-hydroxybenzyl)-5-[(3-cyclopropylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-23 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3URL
 
 | | Endothiapepsin-DB6 complex. | | Descriptor: | DB6 peptide, Endothiapepsin, SULFATE ION | | Authors: | Bailey, D, Sanz-Aparicio, J, Albert, A, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
4AA9
 
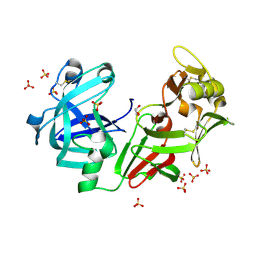 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WZ6
 
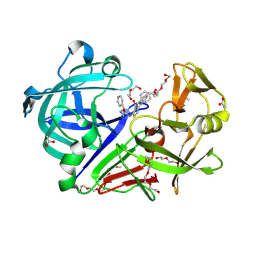 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (5) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3PSY
 
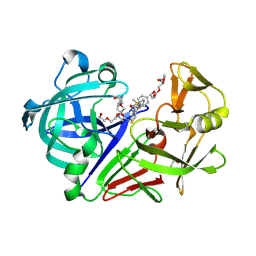 | | Endothiapepsin in complex with an inhibitor based on the Gewald reaction | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3PVK
 
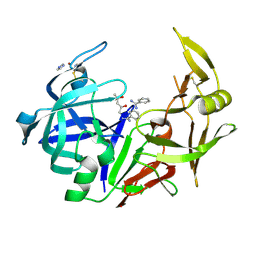 | | Secreted aspartic protease 2 in complex with benzamidine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZAMIDINE, Candidapepsin-2, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-07 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
