4WCA
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230Q, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WCO
 
 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
4WCB
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H309Q | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDE
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T311A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
5DV3
 
 | |
1IXN
 
 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
4WDA
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation P296G, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WC9
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235L | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4FLN
 
 | | Crystal structure of plant protease Deg2 | | Descriptor: | Protease Do-like 2, chloroplastic, Unknown peptide | | Authors: | Gong, W, Liu, L, Sun, R, Gao, F. | | Deposit date: | 2012-06-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
4WFR
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
7ENV
 
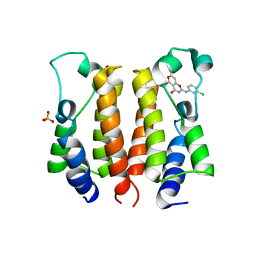 | | crystal structure of NS5 in complex with the N-terminal bromodomain of BRD2 (BRD2-BD1). | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7EO5
 
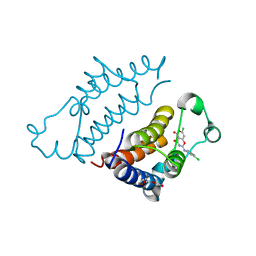 | | Crystal structure of pyrano 1,3, oxazine derivative in complex with the second bromodomain of BRD2 | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5OH4
 
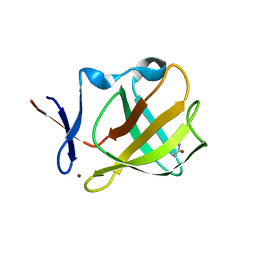 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
7YPU
 
 | | OrfE-CoA-glycylthricin complex | | Descriptor: | Acetyltransferase, COENZYME A, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7YPV
 
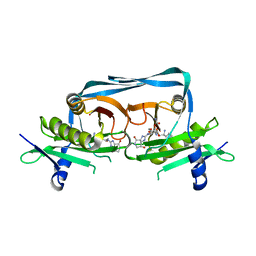 | | Crystal structure of OrE-ST-F | | Descriptor: | Acetyltransferase, Streptothricin F | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
3RDC
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
8CAY
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Siragu, S. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6U
 
 | | PBP AccA-G145YG440Q mutant from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH3
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C75
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 3 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CJU
 
 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKO
 
 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CAW
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with agrocin84 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, [(2R,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
