9BEM
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with seven Tungstates bound | | Descriptor: | Molybdenum-pterin-binding protein, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
8PMK
 
 | |
9BEB
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with eight Tungstates bound | | Descriptor: | Molybdenum-pterin binding domain-containing protein, TUNGSTATE(VI)ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
9BJF
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum in apo form | | Descriptor: | Molybdenum-pterin binding domain-containing protein, SULFATE ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
9BKA
 
 | |
9BKB
 
 | |
9BJV
 
 | |
9BK4
 
 | |
9BKH
 
 | |
5QBW
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-{[2-chloro-5-(2-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-3-oxo-2,3-dihydropyridazin-4-yl)phenyl]methyl}-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
9BED
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with eight molybdates bound | | Descriptor: | MOLYBDATE ION, Molybdenum-pterin-binding protein | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
9BK1
 
 | |
9BK8
 
 | |
9BJY
 
 | |
9BEL
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with five Tungstates bound | | Descriptor: | Molybdenum-pterin binding domain-containing protein, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
5QCJ
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 5-hydroxy-3-{1-[(2S)-2-hydroxy-3-{5-(methylsulfonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl]piperidin-4-yl}-1H-pyrrolo[3,2-c]pyridin-5-ium, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QBZ
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-{[2-chloro-5-(4-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-3-oxo-3,4-dihydropyrazin-2-yl)phenyl]methyl}-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC8
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{3-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
4ZFP
 
 | |
5QC0
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-(dimethylamino)-1-[4-(2-oxo-2-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethyl)piperidin-1-yl]ethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
6FM1
 
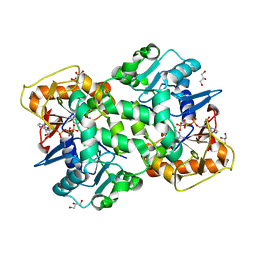 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATPanddGMP at 2.3 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6FCJ
 
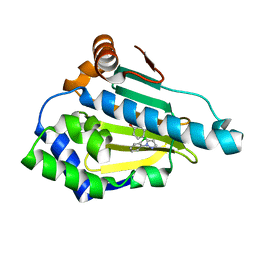 | |
6FCT
 
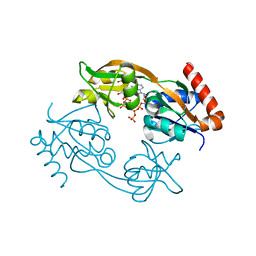 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
6FHP
 
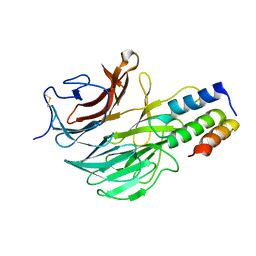 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
