4WPH
 
 | |
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4HN4
 
 | | Tryptophan synthase in complex with alpha aminoacrylate E(A-A) form and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4CL6
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N-(4- Chlorobenzyl)-3-(2-furyl)-1H-1,2,4-triazol-5-amine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-(4-chlorobenzyl)-5-(furan-2-yl)-1H-1,2,4-triazol-3-amine | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Substrate Mimic Allows High Throughput Assay of the Faba Protein and Consequently the Identification of a Novel Inhibitor of Pseudomonas Aeruginosa Faba.
J.Mol.Biol., 428, 2016
|
|
4EG6
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1325 | | Descriptor: | 4-{4-[(1H-benzimidazol-2-ylmethyl)amino]-6-(2-chloro-4-methoxyphenoxy)pyrimidin-2-yl}piperazin-2-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4WXL
 
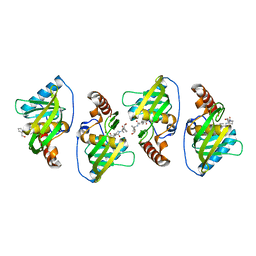 | |
4HMN
 
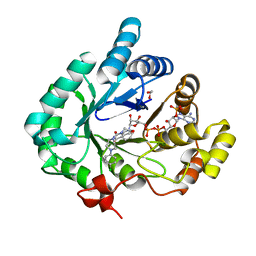 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (4-(4-Chlorophenyl)piperazin-1-yl)(morpholino)methanone (24) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Turnbull, A.P, Flanagan, J.U, Atwell, G.J, Heinrich, D.M, Jamieson, S.M.F, Brooke, D.G, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Morpholylureas are a new class of potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase (AKR1C3).
Bioorg.Med.Chem., 22, 2014
|
|
4EG1
 
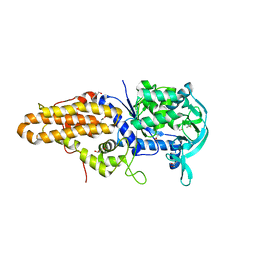 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with substrate Methionine | | Descriptor: | GLYCEROL, METHIONINE, Methionyl-tRNA synthetase, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4X49
 
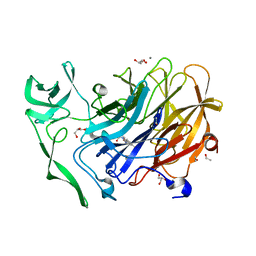 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
3L4S
 
 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 (GAPDH1) from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-12-21 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
4EGA
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1320 | | Descriptor: | 2-({3-[(3,5-dibromo-2-methoxybenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4XO4
 
 | |
2UXF
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
4XN8
 
 | |
4EG3
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with product methionyl-adenylate | | Descriptor: | GLYCEROL, Methionyl-tRNA synthetase, putative, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4I3E
 
 | | Crystal structure of Staphylococcal IMPase - I complexed with products. | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Bhattacharyya, S, Dutta, A, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Staphylococcal IMPase - I complexed with products.
To be Published
|
|
4I3Y
 
 | | Crystal structure of Staphylococcal inositol monophosphatase-1: 100 mM LiCl soaked inhibitory complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
2UZJ
 
 | | Crystal structure of the mature streptococcal cysteine protease, mSpeB | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, STREPTOPAIN | | Authors: | Olsen, J.G, Dagil, R, Niclasen, L.M, Soerensen, O.E, Kragelund, B.B. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the Mature Streptococcal Cysteine Protease Exotoxin Mspeb in its Active Dimeric Form.
J.Mol.Biol., 393, 2009
|
|
4XN2
 
 | |
2UXG
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
4EG5
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1312 | | Descriptor: | 2-({3-[(3,5-dichlorobenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
3KV3
 
 | | Crystal structure of C151S mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH 1)from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GAPDH, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
4XN4
 
 | |
4XN7
 
 | |
