2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
6QBI
 
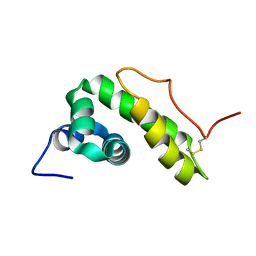 | | NMR structure of BB_P28, Borrelia burgdorferi outer surface lipoprotein | | Descriptor: | Surface protein, mlp lipoprotein family | | Authors: | Fridmanis, J, Otikovs, M, Brangulis, K, Jaudzems, K. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Borrelia burgdorferi outer surface lipoprotein BBP28, a member of the mlp protein family.
Proteins, 2020
|
|
2N3Q
 
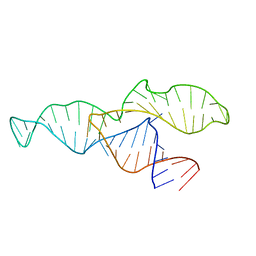 | |
2N3R
 
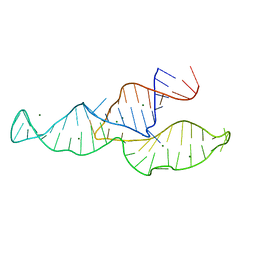 | | NMR structure of the II-III-VI three-way junction from the VS ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement | | Descriptor: | MAGNESIUM ION, RNA (62-MER) | | Authors: | Bonneau, E, Girard, N, Lemieux, S, Legault, P. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Rna, 21, 2015
|
|
2M02
 
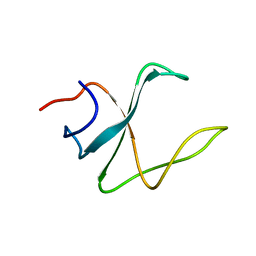 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
7VCK
 
 | |
1FJP
 
 | | NMR Solution Structure of Phospholamban (C41F) | | Descriptor: | CARDIAC PHOSPHOLAMBAN | | Authors: | Lamberth, S, Griesinger, C, Schmid, H, Carafoli, E, Muenchbach, M, Vorherr, T, Krebs, J. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Phospholamban
HELV.CHIM.ACTA, 83, 2000
|
|
1M8L
 
 | |
2MBB
 
 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
5GQS
 
 | | NMR based solution structure of PTS system, galactitol-specific IIB component from methicillin resistant Staphylococcus aureus | | Descriptor: | PTS galactitol transporter subunit IIB | | Authors: | Wahab, A, Schwalbe, H, Shahid, M.F, Richter, C, Jonker, H.R.A. | | Deposit date: | 2016-08-08 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR based solution structure of PTS system, galactitol-specific IIB component from methicillin resistant Staphylococcus aureus
To Be Published
|
|
2LY2
 
 | |
5WOY
 
 | |
5WOX
 
 | |
5WOT
 
 | |
5WOZ
 
 | |
5N14
 
 | |
3BTB
 
 | |
7PS8
 
 | | NMR Structure of the U3 RNA G-quadruplex | | Descriptor: | POTASSIUM ION, RNA (5'-R(*CP*AP*GP*GP*GP*AP*GP*GP*UP*GP*UP*GP*GP*CP*CP*UP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Marquevielle, J, Amrane, S. | | Deposit date: | 2021-09-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the U3 RNA G-quadruplex
To Be Published
|
|
1G6M
 
 | |
1E2B
 
 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
6XYH
 
 | | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2) | | Descriptor: | AMS3 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2)
To Be Published
|
|
6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3)
TO BE PUBLISHED
|
|
2N48
 
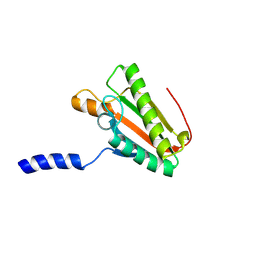 | | EC-NMR Structure of Escherichia coli YiaD Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER553 | | Descriptor: | Probable lipoprotein YiaD | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2XEB
 
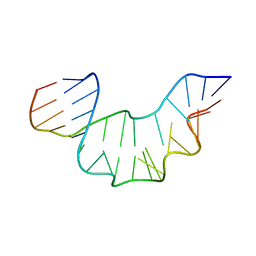 | | NMR STRUCTURE OF THE PROTEIN-UNBOUND SPLICEOSOMAL U4 SNRNA 5' STEM LOOP | | Descriptor: | 5'-R(P*GP*AP*UP*CP*GP*UP*AP*GP*CP*CP*AP*AP*UP*GP*AP* GP*GP*UP*U)-3', 5'-R(P*GP*CP*CP*GP*AP*GP*GP*CP*GP*CP*GP*AP*UP*C)-3' | | Authors: | Falb, M, Amata, I, Gabel, F, Simon, B, Carlomagno, T. | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the K-turn U4 RNA: a combined NMR and SANS study.
Nucleic Acids Res., 38, 2010
|
|
