5IGQ
 
 | |
7M0D
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
5IMA
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F2 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
7LZO
 
 | | DHP B in complex with 2,4-Dibromophenol substrate | | Descriptor: | 2,4-bis(bromanyl)phenol, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic and Structural Studies of 2,4-dihalophenol: Bridging the Functional Gap between Reactivity and Inhibition in Dehaloperoxidase
To Be Published
|
|
5IMF
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F5 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
7LXQ
 
 | |
5IMN
 
 | | Crystal structure of N299A/S303A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Christianson, D.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
5IHY
 
 | |
5VQB
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
5II7
 
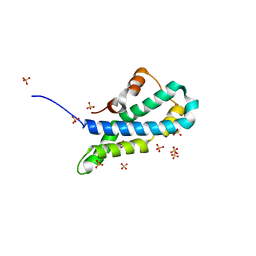 | | In-house sulfur-SAD structure of orthorhombic red abalone lysin at 1.66 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
7LZ5
 
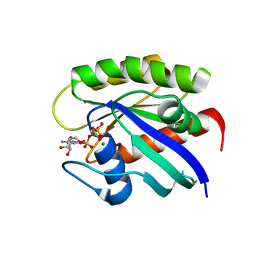 | |
7M0B
 
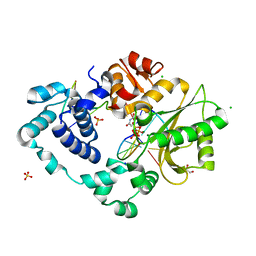 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
5IIN
 
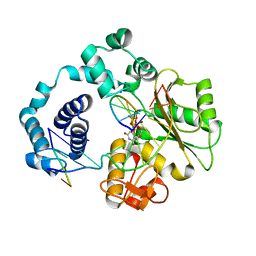 | | Crystal structure of the pre-catalytic ternary extension complex of DNA polymerase lambda with an 8-oxo-dG:dC base-pair | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5VR4
 
 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
7LZR
 
 | |
5IMY
 
 | | Trapped Toxin | | Descriptor: | CD59 glycoprotein, Vaginolysin | | Authors: | Lawrence, S.L, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5VNM
 
 | |
5VO7
 
 | |
7M08
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
7M07
 
 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
1SA4
 
 | | human protein farnesyltransferase complexed with FPP and R115777 | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
5VOX
 
 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5IIV
 
 | | GCN4p pH 1.5 | | Descriptor: | General control protein GCN4 | | Authors: | Brady, M.R, Kaplan, A.R, Alexandrescu, A.T. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structures of GCN4p Are Largely Conserved When Ion Pairs Are Disrupted at Acidic pH but Show a Relaxation of the Coiled Coil Superhelix.
Biochemistry, 56, 2017
|
|
5VRA
 
 | | 2.35-Angstrom In situ Mylar structure of human A2A adenosine receptor at 100 K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Klingel, V, Kuo, A, Kissick, D.J, Ishchenko, A, Lee, M.-Y, Xu, S, Makarov, O, Cherezov, V, Ogata, C.M, Ernst, O.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
7LZN
 
 | | DHP B in complex with 2,4-Dichlorophenol substrate | | Descriptor: | 2,4-dichlorophenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic and Structural Studies of 2,4-dihalophenol: Bridging the Functional Gap between Reactivity and Inhibition in Dehaloperoxidase
To Be Published
|
|
