5TQF
 
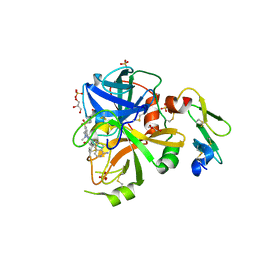 | | Factor VIIa in complex with the inhibitor (11R)-11-[(1-aminoisoquinolin-6-yl)amino]-16-(cyclopropylsulfonyl)-13-methyl-2,13-diazatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaene-3,12-dione | | Descriptor: | (11R)-11-[(1-aminoisoquinolin-6-yl)amino]-16-(cyclopropylsulfonyl)-13-methyl-2,13-diazatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaene-3,12-dione, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2016-10-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Novel Meta-Linked Phenylglycine Macrocyclic FVIIa Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5QEY
 
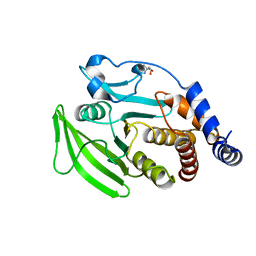 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000708a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[5-(methylsulfanyl)-1,3,4-thiadiazol-2-yl]furan-2-carboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFP
 
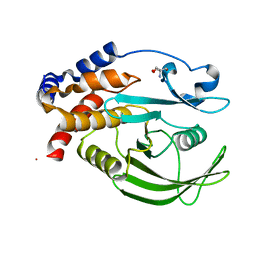 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QG9
 
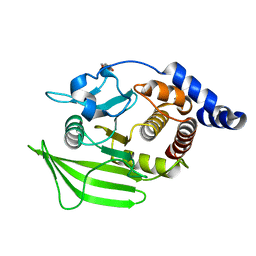 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000595a | | Descriptor: | (1S,3S,4R)-3-[(1S)-1-hydroxypropyl]-2-(methylsulfonyl)-2-azabicyclo[2.2.2]octan-4-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
7VXU
 
 | | Matrix arm of deactive state CI from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-13 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3PX1
 
 | | Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Strontium | | Descriptor: | Calcium-binding protein, STRONTIUM ION | | Authors: | Kumar, S, Kumar, S, Ahmad, E, Khan, R.H, Gourinath, S. | | Deposit date: | 2010-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Flexibility and plasticity of EF-hand motifs: Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Pb2+, Ba2+, and Sr2+.
To be Published
|
|
3PK1
 
 | | Crystal structure of Mcl-1 in complex with the BaxBH3 domain | | Descriptor: | Apoptosis regulator BAX, CADMIUM ION, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2010-11-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis
J.Biol.Chem., 286, 2011
|
|
6KUZ
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | Descriptor: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
5QDM
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000074a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1VZW
 
 | | Crystal structure of the bifunctional protein Pria | | Descriptor: | GLYCEROL, PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Kuper, J, Wilmanns, M. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two-Fold Repeated (Beta-Alpha)(4) Half-Barrels May Provide a Molecular Tool for Dual Substrate Specificity
Embo Rep., 6, 2005
|
|
4IQM
 
 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | tRNA pseudouridine synthase A, mitochondrial | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4DAE
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 6-chloroguanosine | | Descriptor: | 6-chloro-9-(beta-D-ribofuranosyl)-9H-purin-2-amine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
5QF6
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA000281b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methyl-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridine-3-carbonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFN
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000324a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
3PGQ
 
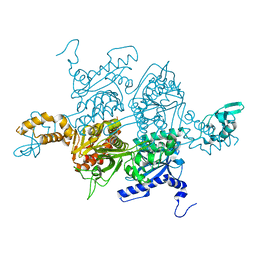 | | Crystal Structure of the Carboxyltransferase Domain of S. cerevisiae Acetyl CoA Carboxylase in Complex with Pinoxaden | | Descriptor: | 8-(2-ethenyl-6-ethyl-4-methylphenyl)tetrahydro-7H-pyrazolo[1,2-d][1,4,5]oxadiazepine-7,9(8H)-dione, Acetyl-CoA carboxylase | | Authors: | Tong, L, Yu, L.P.C, Kim, Y.S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4RBP
 
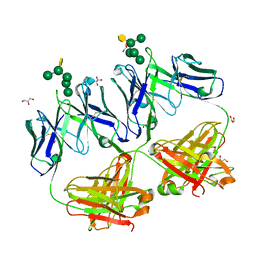 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
6KWY
 
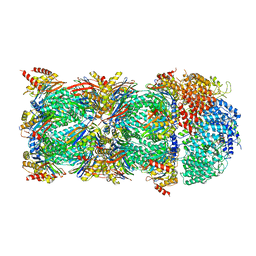 | | human PA200-20S complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
3PIX
 
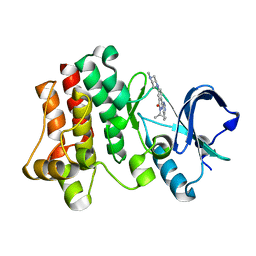 | |
1VK2
 
 | |
5QSW
 
 | | PanDDA analysis group deposition -- Crystal Structure of human STAG1 in complex with Z2856434884 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, Cohesin subunit SA-1 | | Authors: | Newman, J.A, Katis, V.L, Gavard, A.E, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3AXK
 
 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
4LKJ
 
 | | The structure of hemagglutinin L226Q mutant (H3 numbering) from a avian-origin H7N9 influenza virus (A/Anhui/1/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-07-07 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses
Science, 342, 2013
|
|
3PMU
 
 | | Endothiapepsin in complex with a fragment | | Descriptor: | (2S)-1-[(2-fluorobenzyl)oxy]-3-(pyrrolidin-1-yl)propan-2-ol, Endothiapepsin | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-18 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
