2O0T
 
 | |
6FET
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1439 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-[3,4-bis(difluoromethoxy)phenyl]-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)-4,4-dimethylpiperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1439
To be published
|
|
1KAP
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF | | Descriptor: | ALKALINE PROTEASE, CALCIUM ION, TETRAPEPTIDE (GLY SER ASN SER), ... | | Authors: | Baumann, U, Wu, S, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1995-06-08 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif.
EMBO J., 12, 1993
|
|
1NW8
 
 | | Structure of L72P mutant beta class N6-adenine DNA methyltransferase RsrI | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1KPF
 
 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1KPB
 
 | | PKCI-1-APO | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPA
 
 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4JBH
 
 | | 2.2A resolution structure of cobalt and zinc bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
1YD7
 
 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
2C4U
 
 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
5E6C
 
 | | Glucocorticoid receptor DNA binding domain - CCL2 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
7U3W
 
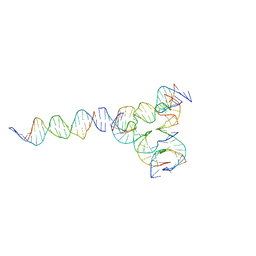 | | [L224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.33 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Z
 
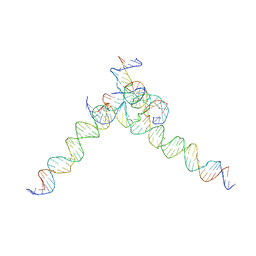 | | [F244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
2BV4
 
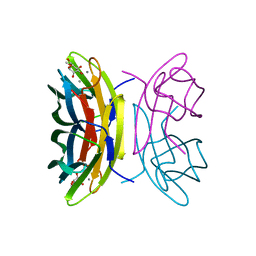 | | 1.0A Structure of Chromobacterium Violaceum Lectin in Complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, LECTIN CV-IIL, methyl alpha-D-mannopyranoside | | Authors: | Pokorna, M, Cioci, G, Perret, S, Rebuffet, E, Adam, J, Gilboa-Garber, N, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2005-06-22 | | Release date: | 2006-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Unusual Entropy Driven Affinity of Chromobacterium Violaceum Lectin Cv-Iil Towards Fucose and Mannose
Biochemistry, 45, 2006
|
|
7U43
 
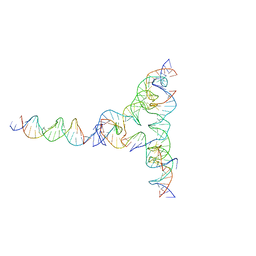 | | [L334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(*TP*AP*CP*AP*CP*CP*GP*AP*TP*CP*AP*CP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U45
 
 | | [L344] Self-assembling tensegrity triangle with three turns, four turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (8.05 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
4JAD
 
 | |
3RFY
 
 | |
5R7Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1220452176 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4N2E
 
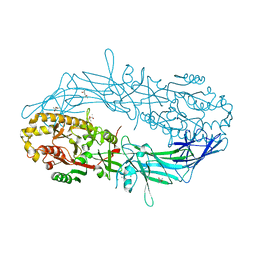 | | Crystal structure of Protein Arginine Deiminase 2 (D123N, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5R80
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z18197050 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl 4-sulfamoylbenzoate | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5E69
 
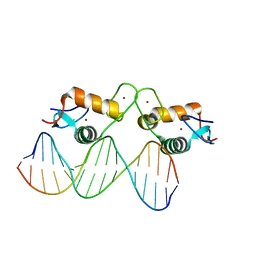 | | Glucocorticoid receptor DNA binding domain - IL8 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*CP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4N2A
 
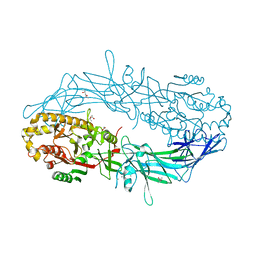 | | Crystal structure of Protein Arginine Deiminase 2 (5 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5E6D
 
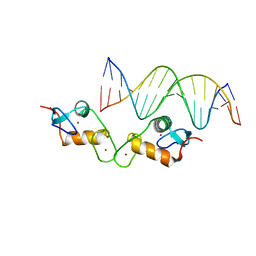 | | Glucocorticoid receptor DNA binding domain - ICAM1 NF-kB response element complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*GP*GP*AP*GP*C)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
