1X44
 
 | | Solution structure of the third ig-like domain of Myosin-dinding protein C, slow-type | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Qin, X.-R, Kurosaki, C, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third ig-like domain of Myosin-dinding protein C, slow-type
to be published
|
|
7YVD
 
 | |
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1ANA
 
 | |
3EBN
 
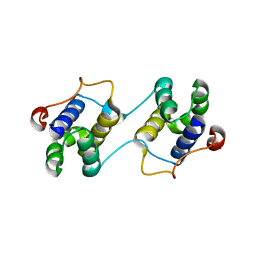 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | Deposit date: | 2008-08-28 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
1A93
 
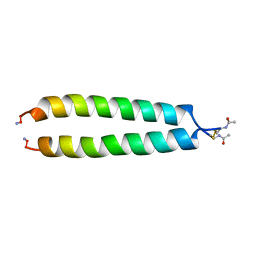 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-04-15 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
4PTD
 
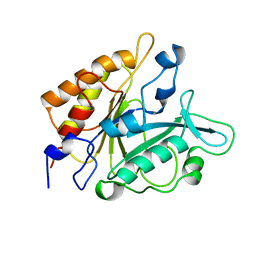 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D274N | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
5EQR
 
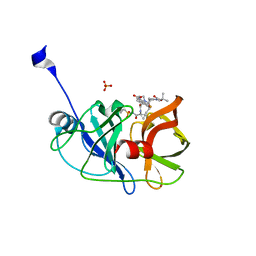 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
4GB1
 
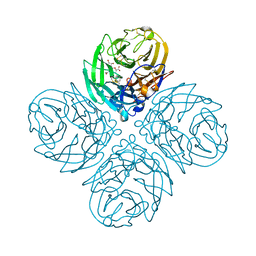 | | Synthesis and Evaluation of Novel 3-C-alkylated-Neu5Ac2en Derivatives as Probes of Influenza Virus Sialidase 150-loop flexibility | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-phenylprop-2-en-1-yl]-D-glycero-L-altro-non-2-enonic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Rudrawar, S, Rameix-Welti, M.-A, Maggioni, A, Dyason, J.C, Rose, F.J, van der Werf, S, Thomson, R.J, Naffakh, N, Russell, R.J.M, von Itzstein, M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility.
Org.Biomol.Chem., 10, 2012
|
|
7D5L
 
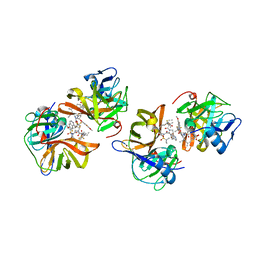 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
1FMK
 
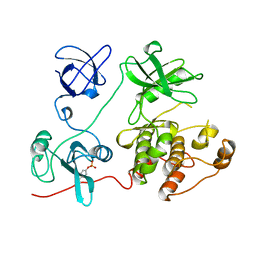 | |
2XXD
 
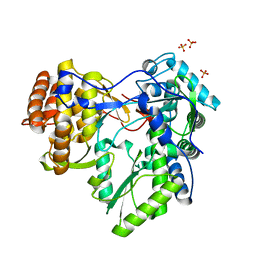 | | HCV-JFH1 NS5B polymerase structure at 1.9 angstrom | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
5ESB
 
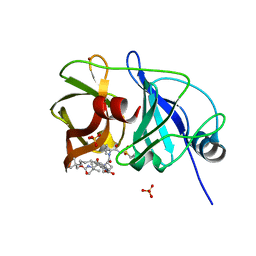 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Y3B
 
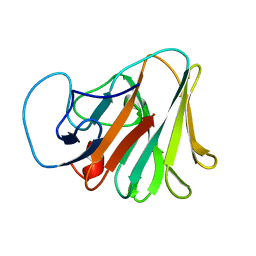 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
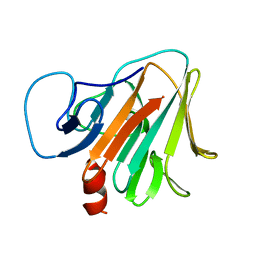 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MWB
 
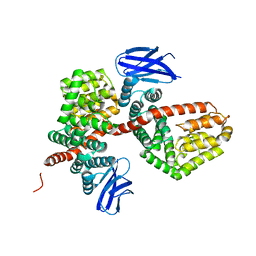 | | ERAP1 binds peptide C-terminus of a SPF sequence (FKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,SPF Sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
7MWC
 
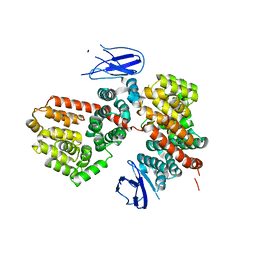 | | ERAP1 binds peptide C-terminus of a LPF sequence (AAAAFKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,LPF sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
9EXV
 
 | |
1URF
 
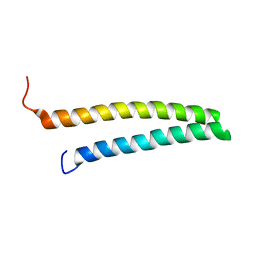 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
4OGP
 
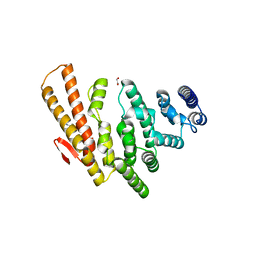 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
1C4L
 
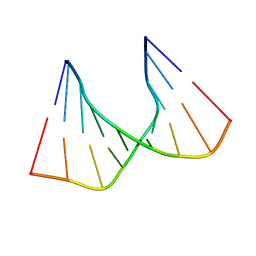 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
4OBC
 
 | | Crystal structure of HCV polymerase NS5b genotype 2a JFH-1 isolate with the S15G, C223H, V321I resistance mutations against the guanosine analog GS-0938 (PSI-3529238) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Abendroth, J, Appleby, T.C. | | Deposit date: | 2014-01-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
2CE1
 
 | | Structure of reduced Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-02 | | Release date: | 2006-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
1BZU
 
 | |
