8PGZ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3528 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-oxidanylidene-1~{H}-pyridazin-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3528
To Be Published
|
|
8PGL
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3027 | | Descriptor: | 7-[(1~{S})-1-[5-(carbamimidamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3027
To Be Published
|
|
8R5U
 
 | |
8R5T
 
 | |
8CGL
 
 | |
5U8O
 
 | |
8T1R
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
2I7T
 
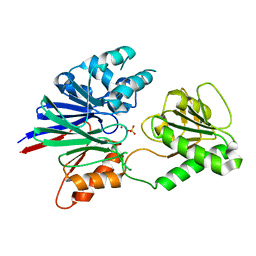 | | Structure of human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
2I7V
 
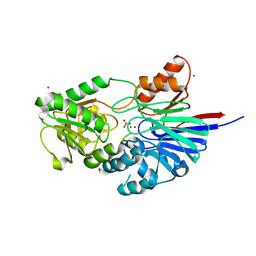 | | Structure of Human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
1QH3
 
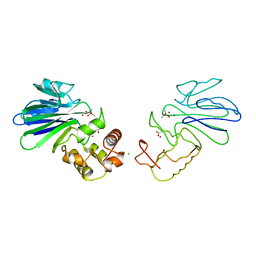 | | HUMAN GLYOXALASE II WITH CACODYLATE AND ACETATE IONS PRESENT IN THE ACTIVE SITE | | Descriptor: | ACETATE ION, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-10 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
1QH5
 
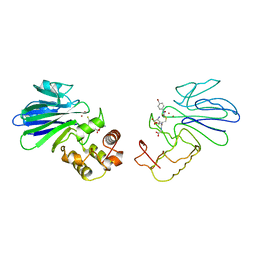 | | HUMAN GLYOXALASE II WITH S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE | | Descriptor: | GLUTATHIONE, PROTEIN (HYDROXYACYLGLUTATHIONE HYDROLASE), S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-11 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
8EWO
 
 | | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, PA1813, ... | | Authors: | Stogios, P.J, Skarina, T, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa
To Be Published
|
|
6I1D
 
 | | Structure of the Ysh1-Mpe1 nuclease complex from S.cerevisiae | | Descriptor: | Endoribonuclease YSH1, GLYCEROL, Protein MPE1, ... | | Authors: | Hill, C.H, Boreikaite, V, Kumar, A, Casanal, A, Kubik, P, Degliesposti, G, Maslen, S, Mariani, A, von Loeffelholz, O, Girbig, M, Skehel, M, Passmore, L.A. | | Deposit date: | 2018-10-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex.
Mol.Cell, 73, 2019
|
|
7PCR
 
 | | Helicobacter pylori RNase J | | Descriptor: | Ribonuclease J | | Authors: | Luisi, B.F, Pei, X.Y. | | Deposit date: | 2021-08-03 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Acetylation regulates the oligomerization state and activity of RNase J, the Helicobacter pylori major ribonuclease.
Nat Commun, 14, 2023
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
4XWW
 
 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6RZ0
 
 | |
4XWT
 
 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6S0I
 
 | |
4NUR
 
 | | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9 | | Descriptor: | MAGNESIUM ION, PSdsA, ZINC ION | | Authors: | Sun, L.F, Su, Y.T, Zhao, Y.H, Cai, Z.X, Wu, Y. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9
To be Published
|
|
6K6W
 
 | |
6K6S
 
 | |
5I0P
 
 | |
1XM8
 
 | | X-RAY STRUCTURE OF GLYOXALASE II FROM ARABIDOPSIS THALIANA GENE AT2G31350 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural studies on a mitochondrial glyoxalase II.
J.Biol.Chem., 280, 2005
|
|
