7XQ3
 
 | |
7XPW
 
 | | Structure of OhTRP14 | | Descriptor: | Thioredoxin domain-containing protein 17 | | Authors: | Wang, S.Q, Huang, S.Q. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into the redox regulation of Oncomelania hupensis TRP14 and its potential role in the snail host response to parasite invasion.
Fish Shellfish Immunol., 128, 2022
|
|
6CXD
 
 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | Descriptor: | Peptidase B, SULFATE ION | | Authors: | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
9DC0
 
 | |
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
6XYV
 
 | |
6ZK0
 
 | | 1.47A human IMPase with ebselen | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | Authors: | Bax, B.D, Fenn, G.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallization and structure of ebselen bound to Cys141 of human inositol monophosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8D8M
 
 | |
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
7VUM
 
 | | Crystal structure of SSB complexed with que | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity.
Int J Mol Sci, 23, 2022
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
8EE2
 
 | |
7YM1
 
 | | Structure of SsbA protein in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Yang, P.C, Chiang, W.Y, Lin, E.S, Huang, C.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil.
Int J Mol Sci, 24, 2023
|
|
5UMO
 
 | |
9PDD
 
 | | 22bin20S complex (NSF-alphaSNAP-2:2 syntaxin-1a:SNAP-25), hydrolyzing, class 29 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Brunger, A.T. | | Deposit date: | 2025-06-30 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural remodeling of target-SNARE protein complexes by NSF enables synaptic transmission.
Nat Commun, 16, 2025
|
|
4Y3E
 
 | | Endothiapepsin in complex with fragment 5 | | Descriptor: | 1H-isoindol-3-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6LXG
 
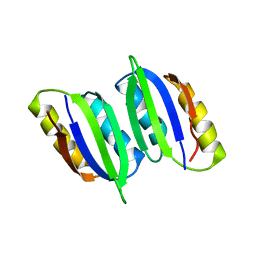 | | NMR solution structure of regulatory ACT domain of the Mycobacterium tuberculosis Rel protein | | Descriptor: | GTP pyrophosphokinase | | Authors: | Shin, J, Singal, B, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Febs J., 288, 2021
|
|
8WQ9
 
 | |
6MVH
 
 | |
6MVF
 
 | |
6MVG
 
 | |
5WT4
 
 | | L-Cysteine-PLP intermediate of NifS from Helicobacter pylori | | Descriptor: | Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Fujishiro, T, Nakamura, R, Takahashi, T. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
6GKF
 
 | |
6KFD
 
 | | Hydroxynitrile lyase from the millipede, Chamberlinius hualienensis, complexed with iodoacetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hydroxynitrile lyase, iodoacetic acid | | Authors: | Motojima, F, Izumi, A, Asano, Y. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | R-hydroxynitrile lyase from the cyanogenic millipede, Chamberlinius hualienensis-A new entry to the carrier protein family Lipocalines.
Febs J., 288, 2021
|
|
5J78
 
 | | Crystal structure of an Acetylating Aldehyde Dehydrogenase from Geobacillus thermoglucosidasius | | Descriptor: | ACETATE ION, Acetaldehyde dehydrogenase (Acetylating), GLYCEROL, ... | | Authors: | Crennell, S.J, Extance, J.P, Danson, M.J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius.
Protein Sci., 25, 2016
|
|
