1GYD
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
5QDG
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000294a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-[(dimethylamino)methyl]-4-methyl-7-oxidanyl-chromen-2-one, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QDO
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOCR000171b | | Descriptor: | 2-(thiophen-2-yl)-1H-imidazole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1GWI
 
 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
6EM5
 
 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6E0W
 
 | | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid | | Descriptor: | 1,2-ETHANEDIOL, 4,6-O-[(1R)-1-carboxyethylidene]-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-alpha-D-galactopyranose-(1-3)-alpha-L-fucopyranose-(1-4)-alpha-L-fucopyranose-(1-3)-beta-D-glucopyranose, Bacteriophage Phi92 gp150, ... | | Authors: | Plattner, M, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Leiman, P.G, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
2CK3
 
 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5QFB
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_PKOOA000283c | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-ethylthieno[2,3-d]pyrimidin-4(3H)-one, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1HAK
 
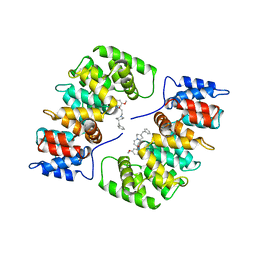 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
1GOY
 
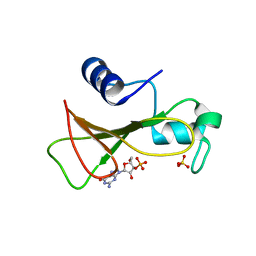 | | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5QG5
 
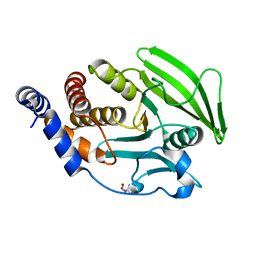 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA000811b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-hydroxybenzonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QOL
 
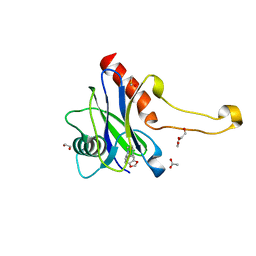 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000435a | | Descriptor: | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1HDI
 
 | | Pig muscle 3-PHOSPHOGLYCERATE KINASE complexed with 3-PG and MgADP. | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Szilagyi, A.N, Ghosh, M, Garman, E, Vas, M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-02-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A 1.8 A resolution structure of pig muscle 3-phosphoglycerate kinase with bound MgADP and 3-phosphoglycerate in open conformation: new insight into the role of the nucleotide in domain closure.
J. Mol. Biol., 306, 2001
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
5QPW
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000632a | | Descriptor: | 1-methyl-5-(phenylamino)-1,2-dihydro-3H-pyrazol-3-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
456C
 
 | | CRYSTAL STRUCTURE OF COLLAGENASE-3 (MMP-13) COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
5QIV
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102998 | | Descriptor: | 1,2-ETHANEDIOL, N'-acetyl-2-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetohydrazide, UNKNOWN LIGAND, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
3MZC
 
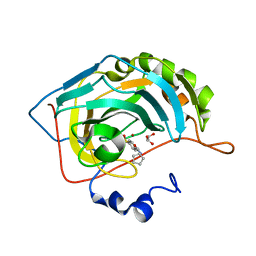 | | Human carbonic ahydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-[(cyclopentylcarbamoyl)amino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
5QJN
 
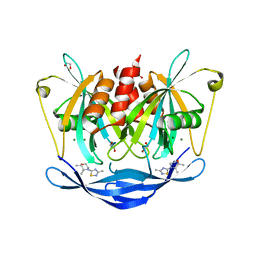 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1439422127 | | Descriptor: | (2S,3R)-2,3-dimethyl-4-(3-methyl-1,2,4-thiadiazol-5-yl)morpholine, 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4FQZ
 
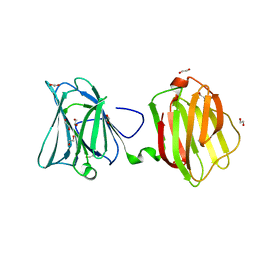 | |
165L
 
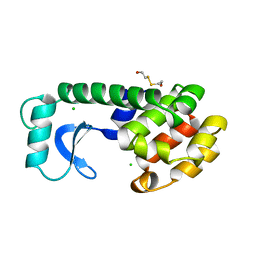 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
7JFY
 
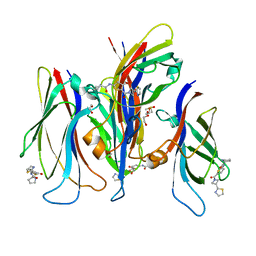 | | GAS41 YEATS domain in complex with 5 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, N-(5-{3-[(2S)-1,3-thiazolidin-2-yl]azetidine-1-carbonyl}thiophen-2-yl)-L-prolinamide, ... | | Authors: | Linhares, B.M, Listunov, D, Winkler, A, Grembecka, J, Cierpicki, T. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.100557 Å) | | Cite: | GAS41 YEATS domain in complex with 5
To Be Published
|
|
3B6T
 
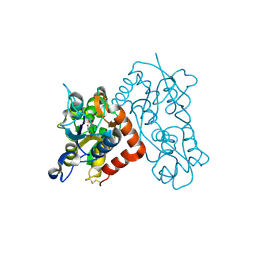 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Cho, Y, Lolis, E, Howe, J.R. | | Deposit date: | 2007-10-29 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
3PWW
 
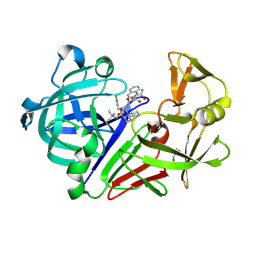 | | Endothiapepsin in complex with saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Endothiapepsin, GLYCEROL | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
