1NBE
 
 | |
1SUE
 
 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
1MUX
 
 | | SOLUTION NMR STRUCTURE OF CALMODULIN/W-7 COMPLEX: THE BASIS OF DIVERSITY IN MOLECULAR RECOGNITION, 30 STRUCTURES | | Descriptor: | CALCIUM ION, CALMODULIN, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE | | Authors: | Osawa, M, Swindells, M.B, Tanikawa, J, Tanaka, T, Mase, T, Furuya, T, Ikura, M. | | Deposit date: | 1997-09-06 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calmodulin-W-7 complex: the basis of diversity in molecular recognition.
J.Mol.Biol., 276, 1998
|
|
1NAT
 
 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
1MPS
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M 197 REPLACED WITH ARG AND TYR M 177 REPLACED WITH PHE (CHAIN M, Y177F, F197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Mcauley-Hecht, K.E, Fyfe, P.K, Ridge, J.P, Prince, S, Hunter, C.N, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1998-03-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies of wild-type and mutant reaction centers from an antenna-deficient strain of Rhodobacter sphaeroides: monitoring the optical properties of the complex from bacterial cell to crystal.
Biochemistry, 37, 1998
|
|
1A9V
 
 | |
1A5R
 
 | | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | | Descriptor: | SUMO-1 | | Authors: | Bayer, P, Arndt, A, Metzger, S, Mahajan, R, Melchior, F, Jaenicke, R, Becker, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the small ubiquitin-related modifier SUMO-1.
J.Mol.Biol., 280, 1998
|
|
1A8N
 
 | | SOLUTION STRUCTURE OF A NA+ CATION STABILIZED DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS FORMED BY G-G-G-C REPEATS OBSERVED IN AAV AND HUMAN CHROMOSOME 19, NMR, 8 STRUCTURES | | Descriptor: | DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS | | Authors: | Kettani, A, Bouaziz, S, Gorin, A, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA.
J.Mol.Biol., 282, 1998
|
|
1A7E
 
 | |
8CGT
 
 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
2RTI
 
 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I222 | | Descriptor: | FORMIC ACID, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTL
 
 | |
1AUQ
 
 | | A1 DOMAIN OF VON WILLEBRAND FACTOR | | Descriptor: | A1 DOMAIN OF VON WILLEBRAND FACTOR, CADMIUM ION | | Authors: | Emsley, J, Cruz, M, Handin, R, Liddington, R. | | Deposit date: | 1997-09-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib.
J.Biol.Chem., 273, 1998
|
|
1AQ2
 
 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tari, L.W, Matte, A, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 1997-08-05 | | Release date: | 1998-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mg(2+)-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions.
Nat.Struct.Biol., 4, 1997
|
|
1AU2
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PROPANONE INHIBITOR | | Descriptor: | 1-[[(4-PHENOXYPHENYL)SULFONYL]AMINO]-3-[[N/N-(4-PYRIDINYLCARBONYL)-L-LEUCYL]AMINO]-2-PROPANOL, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1AV3
 
 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | Descriptor: | RIBONUCLEASE | | Authors: | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | Deposit date: | 1998-05-05 | | Release date: | 1998-10-14 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
1BDW
 
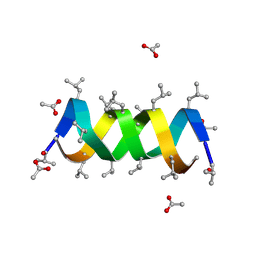 | |
2RTD
 
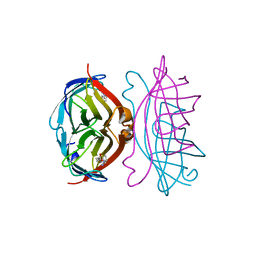 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 1.39, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTR
 
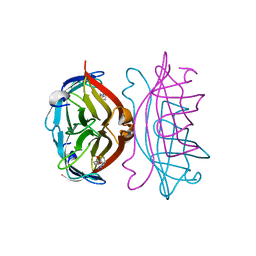 | | STREPTAVIDIN-2-IMINOBIOTIN COMPLEX, PH 4.0, SPACE GROUP I222 | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
1BE6
 
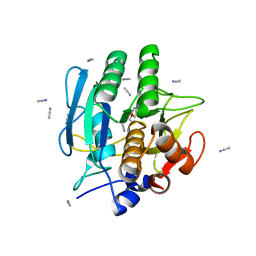 | | TRANS-CINNAMOYL-SUBTILISIN IN ANHYDROUS ACETONITRILE | | Descriptor: | ACETONITRILE, CALCIUM ION, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of x-ray crystal structures of an acyl-enzyme intermediate of subtilisin Carlsberg formed in anhydrous acetonitrile and in water.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BII
 
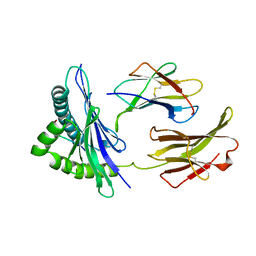 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | Descriptor: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | Authors: | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | Deposit date: | 1998-06-11 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
1BGX
 
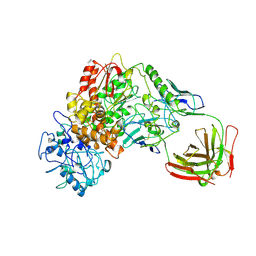 | | TAQ POLYMERASE IN COMPLEX WITH TP7, AN INHIBITORY FAB | | Descriptor: | TAQ DNA POLYMERASE, TP7 MAB | | Authors: | Murali, R, Sharkey, D.J, Daiss, J.L, Krishna Murthy, H.M. | | Deposit date: | 1998-06-02 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BG0
 
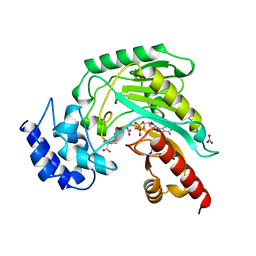 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BDR
 
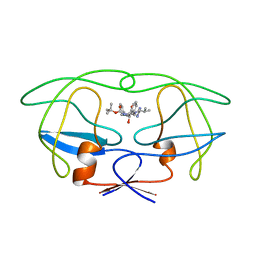 | | HIV-1 (2: 31, 33-37) PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV-1 PROTEASE | | Authors: | Swairjo, M.A, Abdel-Meguid, S.S. | | Deposit date: | 1998-05-10 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural role of the 30's loop in determining the ligand specificity of the human immunodeficiency virus protease.
Biochemistry, 37, 1998
|
|
