2KJN
 
 | |
7BCX
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 8 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7BDM
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 98 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7BD0
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 26 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7BCU
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 1.5 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
5XSI
 
 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KJO
 
 | |
2KSP
 
 | | Mechanism for the selective interaction of C-terminal EH-domain proteins with specific NPF-containing partners | | Descriptor: | CALCIUM ION, EH domain-containing protein 1, MICAL L1 like peptide | | Authors: | Kieken, F, Sharma, M, Jovic, M, Giridharan, S.S, Naslavsky, N, Caplan, S, Sorgen, P.L. | | Deposit date: | 2010-01-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners.
J.Biol.Chem., 285, 2010
|
|
7UDA
 
 | | Structure of the EstG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-03-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
2PBO
 
 | |
2PB1
 
 | | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Hypothetical protein XOG1 | | Authors: | Cutfield, S.M, Cutfield, J.F, Davies, G.J, Sullivan, P.A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A
To be Published
|
|
2L5R
 
 | |
6HL9
 
 | |
6HA5
 
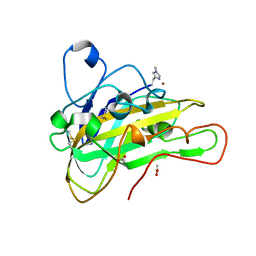 | | AFGH61B L90V/D131S/M134L/A141W VARIANT | | Descriptor: | ACETATE ION, COPPER (II) ION, Endoglucanase, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
3CL4
 
 | | Crystal structure of bovine coronavirus hemagglutinin-esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1PE4
 
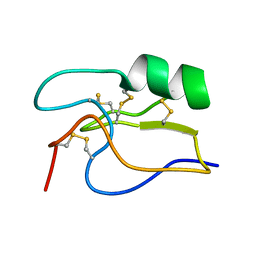 | | SOLUTION STRUCTURE OF TOXIN CN12 FROM CENTRUROIDES NOXIUS ALFA SCORPION TOXIN ACTING ON SODIUM CHANNELS. NMR STRUCTURE | | Descriptor: | Neurotoxin Cn11 | | Authors: | Del Rio-Portilla, F, Hernandez-Marin, E, Pimienta, G, Coronas, F.V, Zamudio, F.Z, Rodrguez de la Vega, R.C, Wanke, E, Possani, L.D. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cn12, a novel peptide from the Mexican scorpion Centruroides noxius with a typical beta-toxin sequence but with alpha-like physiological activity.
Eur.J.Biochem., 271, 2004
|
|
6GPE
 
 | |
6GPN
 
 | |
6HL8
 
 | |
1PTA
 
 | |
6HAQ
 
 | | AFGH61B WILD-TYPE COPPER LOADED | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N, Weihe, C.D. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
2W3Z
 
 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
3CL5
 
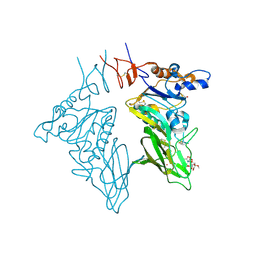 | | Structure of coronavirus hemagglutinin-esterase in complex with 4,9-O-diacetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6H1Z
 
 | | AFGH61B WILD-TYPE | | Descriptor: | ACETATE ION, COPPER (II) ION, Endoglucanase, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
