1WVM
 
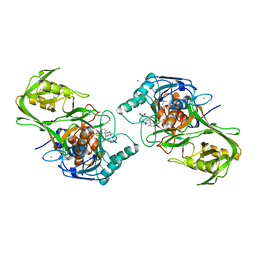 | |
1WMD
 
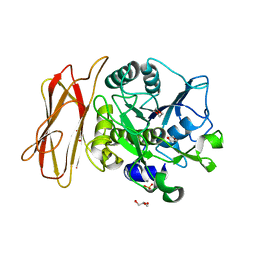 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
5SIC
 
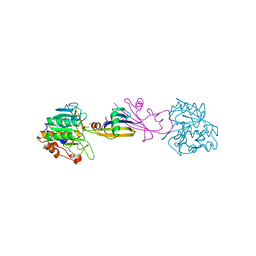 | |
3M0C
 
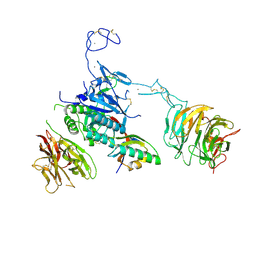 | |
4OV6
 
 | | Crystal structure of PCSK9(53-451) with Adnectin | | Descriptor: | 1,2-ETHANEDIOL, Adnectin, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Khan, J.A. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Pharmacologic Profile of the Adnectin BMS-962476, a Small Protein Biologic Alternative to PCSK9 Antibodies for Low-Density Lipoprotein Lowering.
J.Pharmacol.Exp.Ther., 350, 2014
|
|
3P5C
 
 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
3P5B
 
 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
3QTL
 
 | |
1R0R
 
 | | 1.1 Angstrom Resolution Structure of the Complex Between the Protein Inhibitor, OMTKY3, and the Serine Protease, Subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Ovomucoid, subtilisin carlsberg | | Authors: | Horn, J.R, Ramaswamy, S, Murphy, K.P. | | Deposit date: | 2003-09-22 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and energetics of protein-protein interactions: the role of conformational heterogeneity in OMTKY3 binding to serine proteases
J.Mol.Biol., 331, 2003
|
|
1YU6
 
 | | Crystal Structure of the Subtilisin Carlsberg:OMTKY3 Complex | | Descriptor: | CALCIUM ION, Ovomucoid, Subtilisin Carlsberg | | Authors: | Maynes, J.T, Cherney, M.M, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the subtilisin Carlsberg-OMTKY3 complex reveals two different ovomucoid conformations.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
