2OX5
 
 | | The SoxYZ complex of Paracoccus pantotrophus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Bruno, S, Sauve, V, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The SoxYZ Complex Carries Sulfur Cycle Intermediates on a Peptide Swinging Arm.
J.Biol.Chem., 282, 2007
|
|
2B60
 
 | | Structure of HIV-1 protease mutant bound to Ritonavir | | Descriptor: | GLYCEROL, Gag-Pol polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
3PUO
 
 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
5QHN
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000622a | | Descriptor: | 1,2-ETHANEDIOL, Protein FAM83B, methyl (2~{S},4~{R})-1-(furan-2-ylcarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylate | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3ZW3
 
 | | Fragment based discovery of a novel and selective PI3 Kinase inhibitor | | Descriptor: | N-{6-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]imidazo[1,2-a]pyridin-2-yl}acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Brown, D.G, Hughes, S.J, Milan, D.S, Kilty, I.C, Lewthwaite, R.A, Mathias, J.P, O'Reilly, M.A, Pannifer, A, Phelan, A, Baldock, D.A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment based discovery of a novel and selective PI3 kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
2P3H
 
 | | Crystal structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Uncharacterized CBS domain-containing protein | | Authors: | Cuff, M.E, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin.
TO BE PUBLISHED
|
|
5TBH
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1236 | | Descriptor: | (2~{R})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]amino]-2-azanyl-butanoic acid, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
7HN8
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1318110042 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(but-2-yn-1-yl)(methyl)amino]-1lambda~6~-thiane-1,1-dione, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1QKD
 
 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2VN3
 
 | | Nitrite Reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
7HMJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1578665941 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methyl-1,2,4-thiadiazol-5-yl)-1,4-diazepane, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5CQU
 
 | | Monoclinic Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
7HNS
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z2072621991 | | Descriptor: | 1,2-ETHANEDIOL, 2-(difluoromethoxy)-1-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HOA
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z275165822 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-chlorophenyl)piperazin-1-yl]ethan-1-one, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1W4R
 
 | | Structure of a type II thymidine kinase with bound dTTP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Birringer, M.S, Claus, M.T, Folkers, G, Kloer, D.P, Schulz, G.E, Scapozza, L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-01 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of a Type II Thymidine Kinase with Bound Dttp
FEBS Lett., 579, 2005
|
|
2X1N
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
7HNL
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1251207602 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-methyl-1,3,4-thiadiazol-2-yl)piperidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3A4R
 
 | | The crystal structure of SUMO-like domain 2 in Nip45 | | Descriptor: | 1,2-ETHANEDIOL, NFATC2-interacting protein, SULFATE ION | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
3QHQ
 
 | | Structure of CRISPR-associated protein Csn2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Sag0897 family CRISPR-associated protein | | Authors: | Ellinger, P, Arslan, Z, Wurm, R, Tschapek, B, Pfeffer, K, Wagner, R, Schmitt, L, Pul, U, Smits, S.H. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of CRISPR-associated protein Csn2
To be Published
|
|
5CTG
 
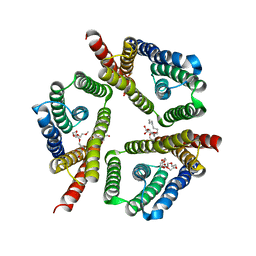 | | The 3.1 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Tao, Y, Perry, K, Feng, L. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
1LAU
 
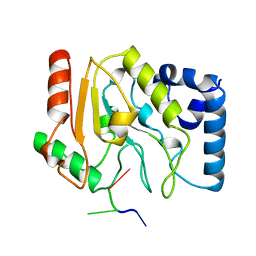 | | URACIL-DNA GLYCOSYLASE | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | Authors: | Pearl, L.H, Savva, R. | | Deposit date: | 1996-01-03 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
5TMT
 
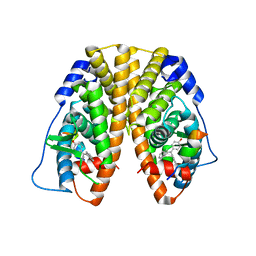 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((1,3-dihydro-2H-inden-2-ylidene)methylene)diphenol | | Descriptor: | 4,4'-[(1,3-dihydro-2H-inden-2-ylidene)methylene]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5QHK
 
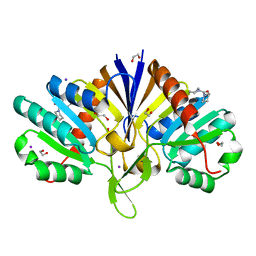 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000010a | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4EZL
 
 | |
5QIU
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0103011 | | Descriptor: | 1,2-ETHANEDIOL, N-{3-[3-(trifluoromethyl)phenyl]prop-2-yn-1-yl}acetamide, UNKNOWN LIGAND, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
