7UFA
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-4-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
7UFC
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-({(2S)-3-phenyl-2-[3-(pyridin-3-yl)propanamido]propyl}sulfanyl)-N-[2-(pyridin-3-yl)ethyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
7UFB
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-({(2S)-3-phenyl-2-[2-(pyridin-3-yl)acetamido]propyl}sulfanyl)-N-[(pyridin-3-yl)methyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
4ABU
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-2 | | Descriptor: | 4-(2,4-dichlorophenoxy)-3-hydroxybenzaldehyde, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
4ABW
 
 | | Crystal Structure of Transthyretin in Complex With Ligand C-6 | | Descriptor: | TRANSTHYRETIN, {4-[4-(hydroxymethyl)-2-methoxyphenoxy]benzene-1,3-diyl}bis[hydroxy(oxo)ammonium] | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
5UTD
 
 | | Sperm whale myoglobin V68A/I107Y with nitrite | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
7Q51
 
 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
 | | human Gid4 bound to a Phe/N-peptide | | Descriptor: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | Authors: | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q4Y
 
 | |
4DIK
 
 | | Flavo Di-iron protein H90A mutant from Thermotoga maritima | | Descriptor: | CHLORIDE ION, FLAVOPROTEIN, MU-OXO-DIIRON | | Authors: | Fang, H, Caranto, J.D, Taylor, A.B, Hart, P.J, Kurtz Jr, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Histidine ligand variants of a flavo-diiron protein: effects on structure and activities.
J.Biol.Inorg.Chem., 17, 2012
|
|
4DIL
 
 | | Flavo Di-iron protein H90N mutant from Thermotoga maritima | | Descriptor: | CHLORIDE ION, FLAVOPROTEIN, MU-OXO-DIIRON | | Authors: | Fang, H, Caranto, J.D, Taylor, A.B, Hart, P.J, Kurtz, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Histidine ligand variants of a flavo-diiron protein: effects on structure and activities.
J.Biol.Inorg.Chem., 17, 2012
|
|
1F63
 
 | | CRYSTAL STRUCTURE OF DEOXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
5UT9
 
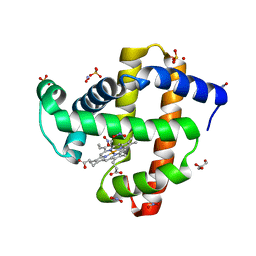 | | Sperm whale myoglobin H64A with nitrite | | Descriptor: | GLYCEROL, Myoglobin, NITRITE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
7M95
 
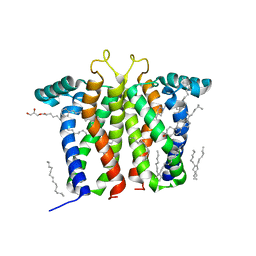 | | Bovine sigma-2 receptor bound to Z1241145220 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[1-(3-phenylpropyl)-1,2,3,6-tetrahydropyridin-4-yl]-1H-pyrrolo[2,3-b]pyridine, CHOLESTEROL, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M96
 
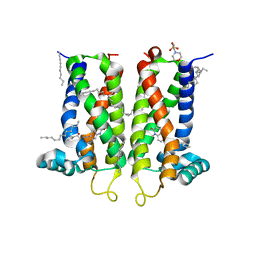 | | Bovine sigma-2 receptor bound to Z4857158944 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-({[(1S)-1-hydroxy-2-methyl-1-phenylpropan-2-yl]amino}methyl)-1-methyl-3,4-dihydroquinolin-2(1H)-one, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
2QA6
 
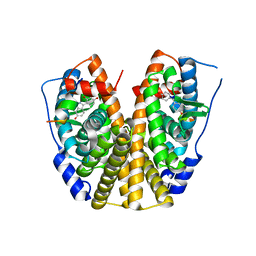 | | Crystal Structure of Estrogen Receptor Alpha mutant 537S Complexed with 4-(6-hydroxy-1H-indazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-(6-HYDROXY-1H-INDAZOL-3-YL)BENZENE-1,3-DIOL, Estrogen receptor, nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
1JOJ
 
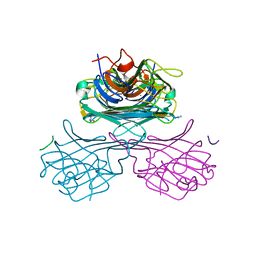 | | CONCANAVALIN A-HEXAPEPTIDE COMPLEX | | Descriptor: | CALCIUM ION, Concanavalin-Br, HEXAPEPTIDE, ... | | Authors: | Jain, D, Kaur, K, Salunke, D.M. | | Deposit date: | 2001-07-30 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced binding of a rationally designed peptide ligand of concanavalin a arises from improved geometrical complementarity.
Biochemistry, 40, 2001
|
|
1JZK
 
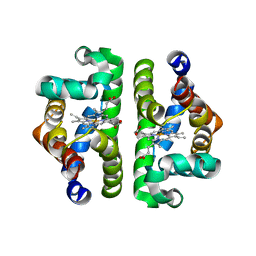 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
2NYJ
 
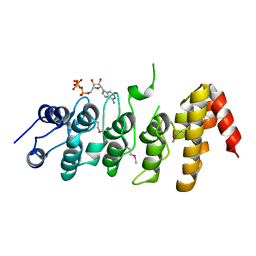 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
5MGR
 
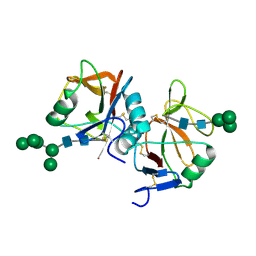 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
4LON
 
 | |
4LUE
 
 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6YUH
 
 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
2DBE
 
 | | CRYSTAL STRUCTURE OF A BERENIL-DODECANUCLEOTIDE COMPLEX: THE ROLE OF WATER IN SEQUENCE-SPECIFIC LIGAND BINDING | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Brown, D.G, Sanderson, M.R, Skelly, J.V, Jenkins, T.C, Brown, T, Garman, E, Stuart, D.I, Neidle, S. | | Deposit date: | 1990-03-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding.
EMBO J., 9, 1990
|
|
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
