6HK7
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C50 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyethyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
4BJM
 
 | | Crystal structure of the flax-rust effector avrM | | Descriptor: | AVRM, CHLORIDE ION | | Authors: | Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the Flax-Rust Effector Avrm Reveal Insights Into the Molecular Basis of Plant-Cell Entry and Effector-Triggered Immunity
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CDB
 
 | | Crystal Structure of V66L CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, SODIUM ION, ... | | Authors: | Capdevila, D.A, Campanello, G, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
6CBC
 
 | |
4BJN
 
 | | Crystal structure of the flax-rust effector AvrM-A | | Descriptor: | AVRM-A | | Authors: | Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Flax-Rust Effector Avrm Reveal Insights Into the Molecular Basis of Plant-Cell Entry and Effector-Triggered Immunity
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6I1J
 
 | |
6E8L
 
 | | Crystal Structure of Alkyl hydroperoxidase D (AhpD) from Streptococcus pneumoniae (Strain D39/ NCTC 7466) | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Meng, Y, Davies, J, North, R, Coombes, D, Horne, C, Hampton, M, Dobson, R. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function analyses of alkylhydroperoxidase D fromStreptococcus pneumoniaereveal an unusual three-cysteine active site architecture.
J.Biol.Chem., 295, 2020
|
|
6K5Q
 
 | | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia | | Descriptor: | ACETATE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Kanal Elamparithi, B, Ankur, T, Sivakumar, M, Gunasekaran, K. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia
To Be Published
|
|
3ZG8
 
 | | Crystal Structure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5N13
 
 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
8V7F
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T*())-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
8V7E
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+
To Be Published
|
|
5NF5
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with CIP-AS at 2.85 A resolution | | Descriptor: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | Deposit date: | 2017-03-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
4AQP
 
 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
5O4F
 
 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with the agonist LM-12b at 2.10 A resolution | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
8SXD
 
 | |
5MA6
 
 | | GFP-binding DARPin 3G124nc | | Descriptor: | 1,2-ETHANEDIOL, 3G124nc, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAK
 
 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA4
 
 | | GFP-binding DARPin fusion gc_K7 | | Descriptor: | Green fluorescent protein, K7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
456C
 
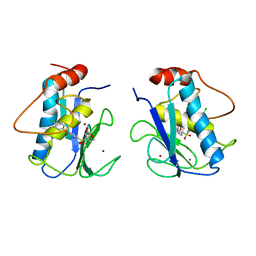 | | CRYSTAL STRUCTURE OF COLLAGENASE-3 (MMP-13) COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
3ZLJ
 
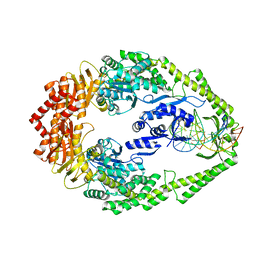 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
8U1W
 
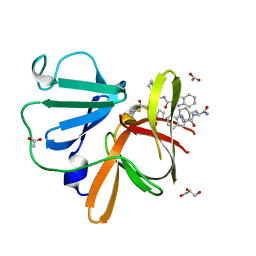 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
8U1V
 
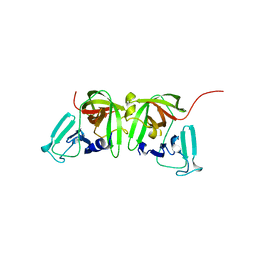 | |
