2BB9
 
 | | Structure of HIV1 protease and AKC4p_133a complex. | | Descriptor: | 2-ETHOXYETHYL (1S,2S)-3-{(2S)-4-[(3AS,8S,8AR)-2-OXO-3,3A,8,8A-TETRAHYDRO-2H-INDENO[1,2-D][1,3]OXAZOL-8-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1PYE
 
 | | Crystal structure of CDK2 with inhibitor | | Descriptor: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | Authors: | Zhang, F, Hamdouchi, C. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
3IV0
 
 | |
2BDU
 
 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
5ET5
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET6
 
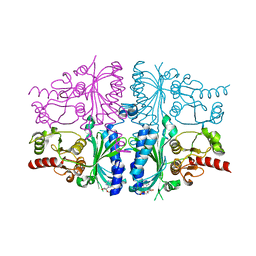 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
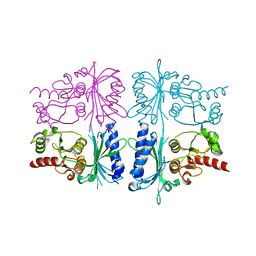 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3OP7
 
 | |
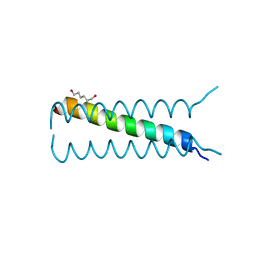
4HLR
 
 | |
2C3N
 
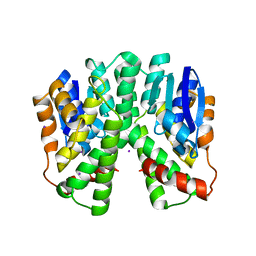 | | Human glutathione-S-transferase T1-1, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
3ING
 
 | |
7ZFG
 
 | | VDR complex with aromatic D-ring analog | | Descriptor: | (1R,3S,5Z)-4-methylidene-5-[(E)-9-methyl-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]-9-oxidanyl-dec-2-enylidene]cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Design, Synthesis, Biological Activity, and Structural Analysis of Novel Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3.
J.Med.Chem., 65, 2022
|
|
3OSD
 
 | |
3IUP
 
 | |
1OM1
 
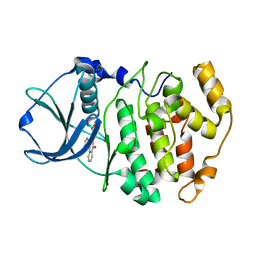 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
3IB5
 
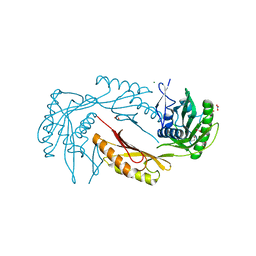 | |
4AA4
 
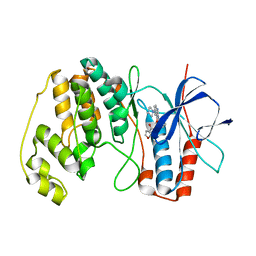 | | P38ALPHA MAP KINASE BOUND TO CMPD 22 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]PHENYL]FURAN-3-CARBOXAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3IOP
 
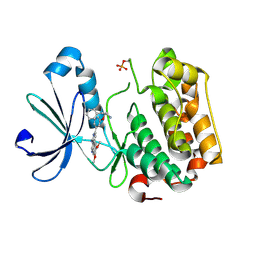 | | PDK-1 in complex with the inhibitor Compound-8i | | Descriptor: | 2-(5-{[(2R)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
5X1W
 
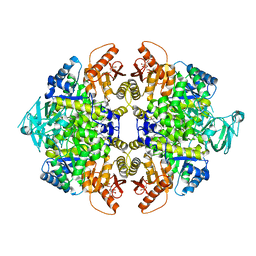 | | PKM2 in complex with compound 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[2,3-bis(chloranyl)phenyl]carbonyl-N-[2-[[4-[2,3-bis(chloranyl)phenyl]carbonyl-1-methyl-pyrrol-2-yl]carbonylamino]ethyl]-1-methyl-pyrrole-2-carboxamide, Pyruvate kinase PKM | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2017-01-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and structure-guided fragment-linking of 4-(2,3-dichlorobenzoyl)-1-methyl-pyrrole-2-carboxamide as a pyruvate kinase M2 activator
Bioorg. Med. Chem., 25, 2017
|
|
3EGE
 
 | |
3QGH
 
 | |
3CGG
 
 | |
3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3LIN
 
 | | crystal structure of HTLV protease complexed with the inhibitor, KNI-10562 | | Descriptor: | N-[(2S,3S)-4-{(4R)-4-[(2,2-dimethylpropyl)carbamoyl]-5,5-dimethyl-1,3-thiazolidin-3-yl}-3-hydroxy-4-oxo-1-phenylbutan-2-yl]-N~2~-{(2S)-2-[(methoxycarbonyl)amino]-2-phenylacetyl}-3-methyl-L-valinamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
4PQ9
 
 | |
