6XH3
 
 | |
6XH2
 
 | |
6XH1
 
 | |
6XH0
 
 | |
2F8E
 
 | | Foot and Mouth Disease Virus RNA-dependent RNA polymerase in complex with uridylylated VPg protein | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA-dependent RNA polymerase, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2005-12-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a protein primer-polymerase complex in the initiation of genome replication
Embo J., 25, 2006
|
|
3H3D
 
 | |
4DWQ
 
 | | RNA ligase RtcB-GMP/Mn(2+) complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MALONATE ION, ... | | Authors: | Okada, C, Xia, S, Englert, M, Yao, M, Soll, D, Wang, J. | | Deposit date: | 2012-02-26 | | Release date: | 2012-09-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2CJQ
 
 | |
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
5U0Q
 
 | | RNA-DNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, DNA/RNA (5'-R(*GP*GP*AP*GP*C)-D(P*T)-R(P*A)-3'), MAGNESIUM ION, ... | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-11-26 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | RNA-DNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
4RNP
 
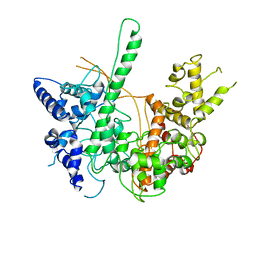 | | BACTERIOPHAGE T7 RNA POLYMERASE, HIGH SALT CRYSTAL FORM, LOW TEMPERATURE DATA, ALPHA-CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Liu, Z.J, Sousa, R, Rose, J.P, Wang, B.C. | | Deposit date: | 1997-09-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution.
Nature, 364, 1993
|
|
8THV
 
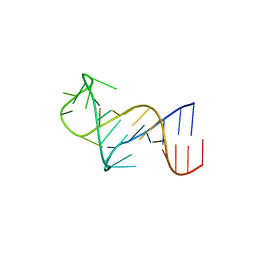 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
5NPA
 
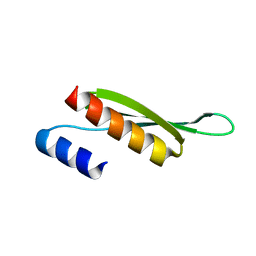 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
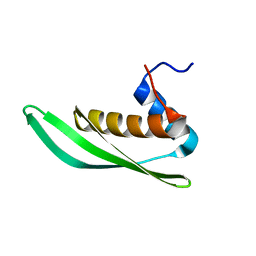 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
3IM2
 
 | |
3IM1
 
 | |
6MJ0
 
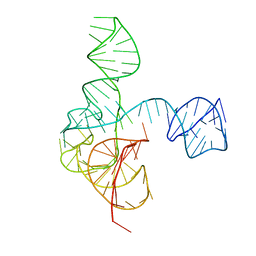 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | Descriptor: | RNA (101-MER) | | Authors: | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
2B7E
 
 | | First FF domain of Prp40 Yeast Protein | | Descriptor: | Pre-mRNA processing protein PRP40 | | Authors: | Gasch, A, Wiesner, S, Martin-Malpartida, P, Ramirez-Espain, X, Ruiz, L, Macias, M.J. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains.
J.Biol.Chem., 281, 2006
|
|
5UZZ
 
 | |
5V1L
 
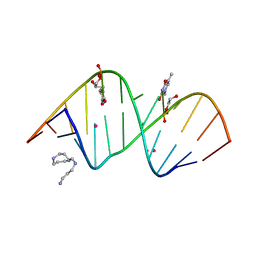 | | Structure of S-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*(ZTH)P*AP*GP*CP*G)-3'), SPERMINE, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
5V1K
 
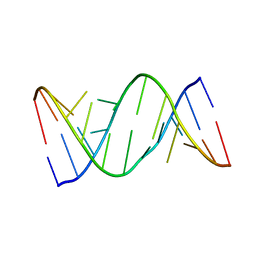 | | Structure of R-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*(5BU)P*(8RJ)P*AP*GP*CP*G)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 2017
|
|
3SBG
 
 | |
2NOQ
 
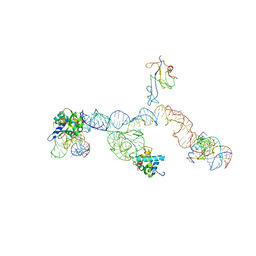 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8PXS
 
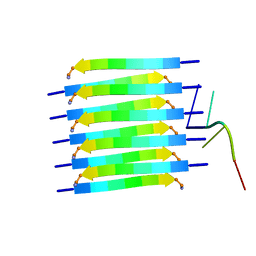 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
