6EQM
 
 | | Crystal Structure of Human BACE-1 in Complex with CNP520 | | Descriptor: | Beta-secretase 1, ~{N}-[6-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-5-fluoranyl-pyridin-2-yl]-3-chloranyl-5-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Wirth, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The BACE-1 inhibitor CNP520 for prevention trials in Alzheimer's disease.
EMBO Mol Med, 10, 2018
|
|
6FGY
 
 | | Crystal Structure of Human BACE-1 in Complex with amino-1,4-oxazine compound 4 | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R})-5-azanyl-3-methyl-2,6-dihydro-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of amino-1,4-oxazines as potent BACE-1 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7OT9
 
 | |
7P4U
 
 | |
5G0R
 
 | |
5FLY
 
 | | The FhuD protein from S.pseudintermedius | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Malito, E. | | Deposit date: | 2015-10-29 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8EPN
 
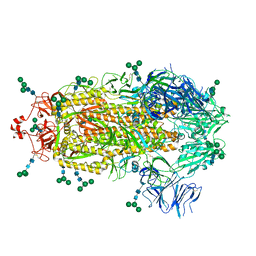 | |
8EPQ
 
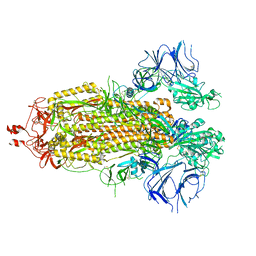 | | Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs exposed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Williams, J.A, Harshbarger, W. | | Deposit date: | 2022-10-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and computational design of a SARS-CoV-2 spike antigen with improved expression and immunogenicity.
Sci Adv, 9, 2023
|
|
8EPP
 
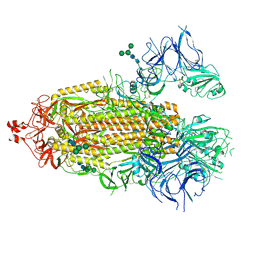 | |
5CDZ
 
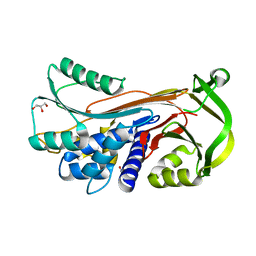 | | Crystal structure of conserpin in the latent state | | Descriptor: | Conserpin in the latent state, GLYCEROL | | Authors: | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
7TZB
 
 | | Crystal structure of the human mitochondrial seryl-tRNA synthetase (mt SerRS) bound with a seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial | | Authors: | Kuhle, B, Hirschi, M, Doerfel, L, Lander, G, Schimmel, P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7U2B
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7U2A
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (38-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
5CDX
 
 | |
5BXZ
 
 | | H17 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | Non-structural protein 1 | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
5BY1
 
 | | H18 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, PENTAETHYLENE GLYCOL | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
4PI0
 
 | |
4PHZ
 
 | |
4PI2
 
 | |
6CUJ
 
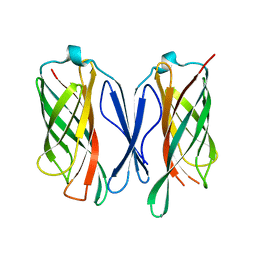 | |
4U9R
 
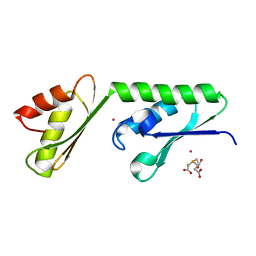 | | Structure of the N-terminal Extension from Cupriavidus metallidurans CzcP | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CADMIUM ION, CzcP cation efflux P1-ATPase | | Authors: | Smith, A.T, Rosenzweig, A.C. | | Deposit date: | 2014-08-06 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A new metal binding domain involved in cadmium, cobalt and zinc transport.
Nat.Chem.Biol., 11, 2015
|
|
7MEI
 
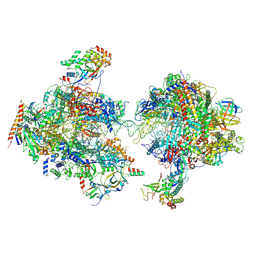 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7NEG
 
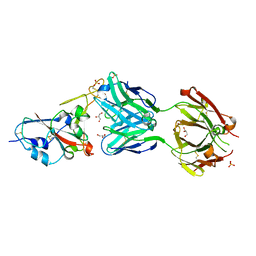 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody COVOX-269 Fab heavy chain, Antibody COVOX-269 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-02-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
7NEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-02-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
7NB6
 
 | |
