8Q36
 
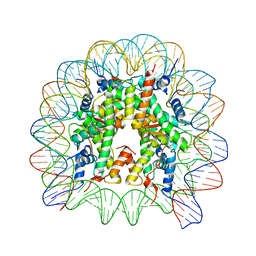 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Foamy Virus GAG Peptide-Au[I] Compound) | | Descriptor: | DNA (145-MER), GAG structural protein, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q3X
 
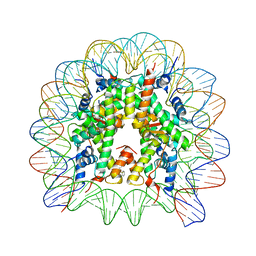 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus LANA Peptide-Au[I] Compound) | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OSL
 
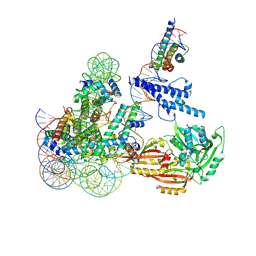 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTS
 
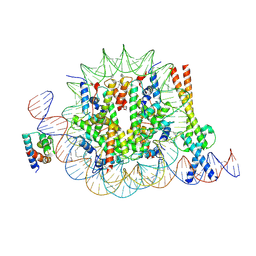 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8VML
 
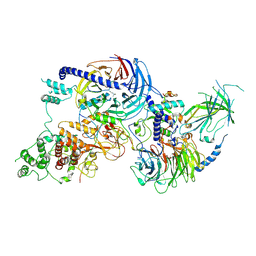 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCY
 
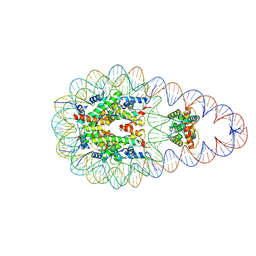 | | Structure of nucleosome complexed with two DEK molecules | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-08 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
8KE0
 
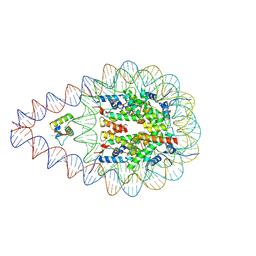 | | Structure of H1.2 bound to the nucleosome | | Descriptor: | DNA (193-MER), Histone H1.2, Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-11 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
8KD1
 
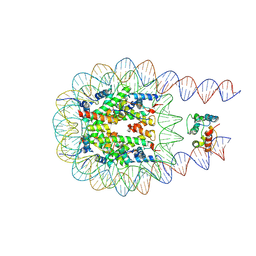 | | Structure of nucleosome complexed with one DEK molecule | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
8F86
 
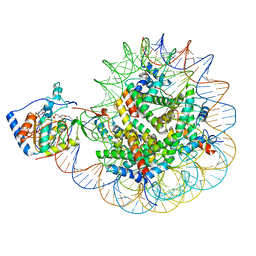 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
6YIH
 
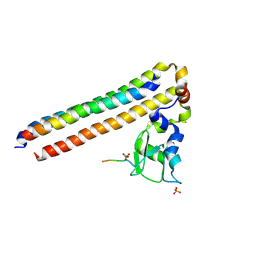 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 2.6 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Histone H3.1, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6XPR
 
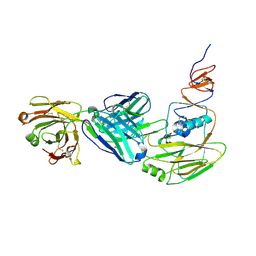 | | Human antibody D2 H1-1/H3-1 H3 in complex with the influenza hemagglutinin head domain of A/Texas/50/2012(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C, Lee, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
9GMR
 
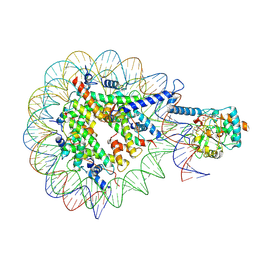 | | SIRT7-H3K36MTUnucleosome complex | | Descriptor: | DNA (149-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
9GMK
 
 | | SIRT7:H3K18DTU nucleosome complex | | Descriptor: | DNA (148-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
8OTT
 
 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8VMJ
 
 | | H3K4me3 nucleosome bound to PRC2_AJ119-450 | | Descriptor: | DNA (157-MER), Histone H2A, Histone H2B, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VOB
 
 | | H3K36me3-modified nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VMN
 
 | | H3K4me3 nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A, Histone H2B, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6UPL
 
 | | Structure of FACT_subnucleosome complex 2 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
