8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VSK
 
 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKD
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
7S86
 
 | |
8T62
 
 | |
8T63
 
 | |
8T61
 
 | |
5Z0A
 
 | | ST0452(Y97N)-GlcNAc binding form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
9FBD
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Thermus thermophilus HB27 complexed to NAD+ | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hurtado-Guerrero, R, Macias-Leon, J, Gines-Alcober, I, Gonzalez-Ramirez, A.M. | | Deposit date: | 2024-05-13 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop engineering of enzymes to control their immobilization and ultimately fabricate more efficient heterogeneous biocatalysts.
Protein Sci., 34, 2025
|
|
6VIF
 
 | | Human LRH-1 ligand-binding domain bound to agonist cpd 15 and fragment of coregulator TIF-2 | | Descriptor: | N-[(8beta,11alpha,12alpha)-8-{[methyl(phenyl)amino]methyl}-1,6:7,14-dicycloprosta-1(6),2,4,7(14)-tetraen-11-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of a new class of liver receptor homolog-1 (LRH-1) agonists by photoredox conjugate addition.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1H22
 
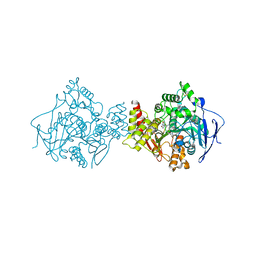 | | Structure of acetylcholinesterase (E.C. 3.1.1.7) complexed with (S,S)-(-)-bis(10)-hupyridone at 2.15A resolution | | Descriptor: | (S,S)-(-)-N,N'-DI-5'-[5',6',7',8'-TETRAHYDRO- 2'(1'H)-QUINOLYNYL]-1,10-DIAMINODECANE DIHYDROCHLORIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.-F, Pang, Y.-P, Silman, I, Sussman, J.L. | | Deposit date: | 2002-07-30 | | Release date: | 2002-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Acetylcholinesterase Complexed with Bivalent Ligands Related to Huperzine A: Experimental Evidence for Species-Dependent Protein-Ligand Complementarity
J.Am.Chem.Soc., 125, 2003
|
|
9FXF
 
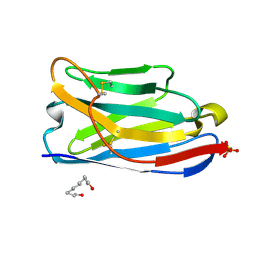 | |
9JS5
 
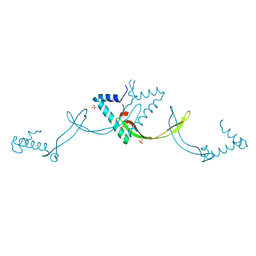 | | Crystal structure of the ASFV-derived histone-like protein pA104R | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Viral histone-like protein | | Authors: | Li, Q, Shao, H, Yi, D, Cen, S. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thonningianin A disrupts pA104R-DNA binding and inhibits African swine fever virus replication.
Emerg Microbes Infect, 14, 2025
|
|
9JUR
 
 | |
8RD6
 
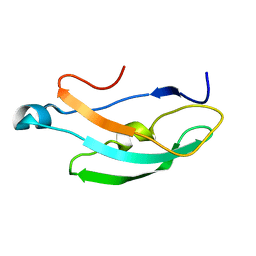 | |
5Z3J
 
 | |
1H23
 
 | | Structure of acetylcholinesterase (E.C. 3.1.1.7) complexed with (S,S)-(-)-bis(12)-hupyridone at 2.15A resolution | | Descriptor: | (S,S)-(-)-N,N'-DI-5'-[5',6',7',8'-TETRAHYDRO- 2'(1'H)-QUINOLYNYL]-1,12-DIAMINODODECANE DIHYDROCHLORIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.F, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2002-07-30 | | Release date: | 2002-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Acetylcholinesterase Complexed with Bivalent Ligands Related to Huperzine A: Experimental Evidence for Species-Dependent Protein-Ligand Complementarity
J.Am.Chem.Soc., 125, 2003
|
|
5AZE
 
 | | Fab fragment of calcium-dependent antigen binding antibody, 6RL#9 | | Descriptor: | 6RL#9 FAB HEAVY CHAIN, 6RL#9 FAB LIGHT CHAIN, CALCIUM ION | | Authors: | Kadono, S, Hironiwa, N, Ishii, S, Igawa, T, Hattori, K. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calcium-dependent antigen binding as a novel modality for antibody recycling by endosomal antigen dissociation
Mabs, 8, 2016
|
|
5Z37
 
 | |
5DQF
 
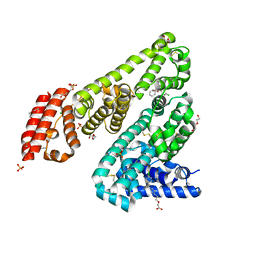 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
4ZSU
 
 | |
7LK3
 
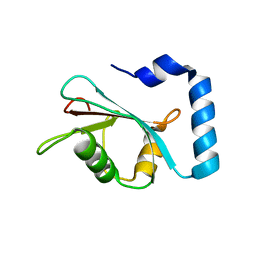 | | Crystal structure of untwinned human GABARAPL2 | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | Authors: | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7RVA
 
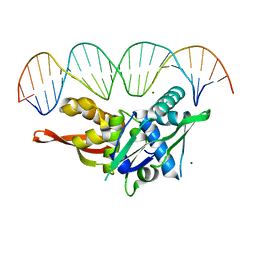 | | Updated Crystal Structure of Replication Initiator Protein REPE54. | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7SGC
 
 | |
5Z3I
 
 | |
