7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
8CNR
 
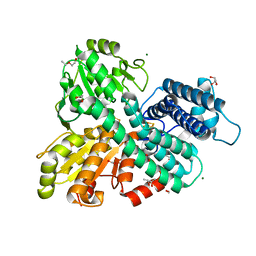 | | Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus as isolated in a reduced state at 1.45-A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
8CKW
 
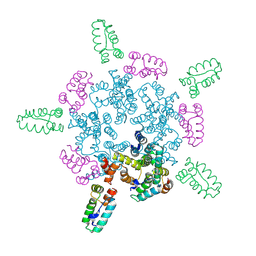 | |
8CNS
 
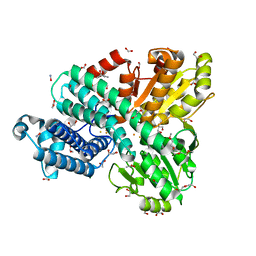 | | The Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus in a mixed redox state after soaking with hydroxylamine, at 1.36-A resolution. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
8CL1
 
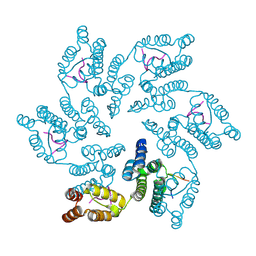 | |
7Z0T
 
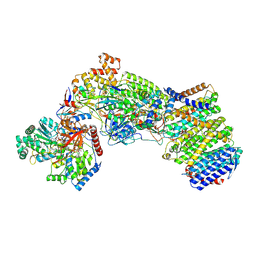 | | Structure of the Escherichia coli formate hydrogenlyase complex (aerobic preparation, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBONMONOXIDE-(DICYANO) IRON, FE (III) ION, ... | | Authors: | Steinhilper, R, Murphy, B.J. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Nat Commun, 13, 2022
|
|
8CKV
 
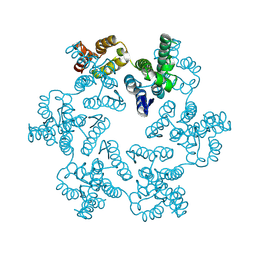 | |
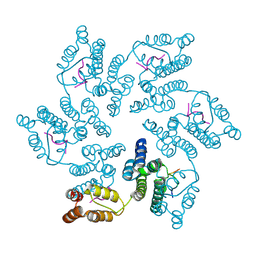
8CL3
 
 | |
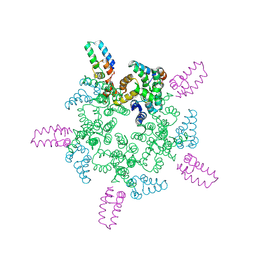
8CL4
 
 | |
8CKZ
 
 | |
8CL0
 
 | |
8CL2
 
 | |
8CKX
 
 | |
8CKY
 
 | |
7Z0S
 
 | |
7QPG
 
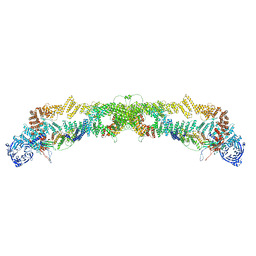 | | Human RZZ kinetochore corona complex. | | Descriptor: | Centromere/kinetochore protein zw10 homolog, Kinetochore-associated protein 1, Protein zwilch homolog | | Authors: | Raisch, T, Ciossani, G, d'Amico, E, Cmetowski, V, Carmignani, S, Maffini, S, Merino, F, Wohlgemuth, S, Vetter, I.R, Raunser, S, Musacchio, A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores.
Embo J., 41, 2022
|
|
7ZQQ
 
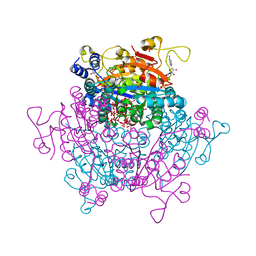 | | Molybdenum storage protein - LB131H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I. | | Deposit date: | 2022-05-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
7R1Y
 
 | |
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
7ZMB
 
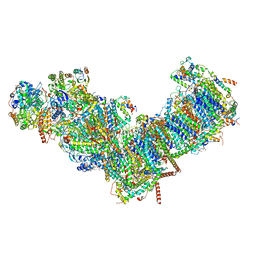 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM8
 
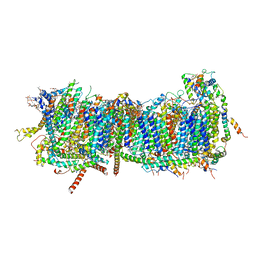 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMH
 
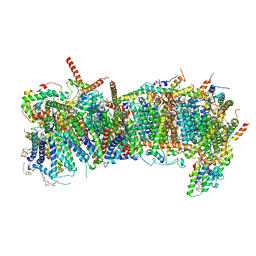 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8CIW
 
 | | Methylsuccinyl-CoA dehydrogenase from Pseudomonas migulae with bound FAD and (2S)-methylsuccinyl-CoA | | Descriptor: | (2S)-Methylsuccinyl-CoA, (2S)-methylsuccinyl-CoA dehydrogenase, CITRATE ANION, ... | | Authors: | Zarzycki, J, McLean, R, Erb, T.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring alternative pathways for the in vitro establishment of the HOPAC cycle for synthetic CO 2 fixation.
Sci Adv, 9, 2023
|
|
7ZM7
 
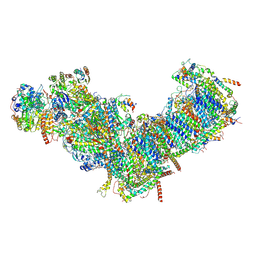 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZME
 
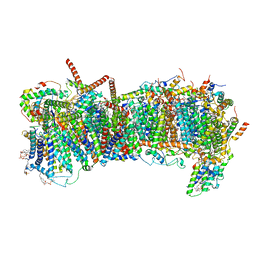 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
