2BG5
 
 | | Crystal Structure of the Phosphoenolpyruvate-binding Enzyme I-Domain from the Thermoanaerobacter tengcongensis PEP: Sugar Phosphotransferase System (PTS) | | Descriptor: | PHOSPHOENOLPYRUVATE-PROTEIN KINASE | | Authors: | Oberholzer, A.E, Bumann, M, Schneider, P, Baechler, C, Siebold, C, Baumann, U, Erni, B. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the Phosphoenolpyruvate-Binding Enzyme I-Domain from the Thermoanaerobacter Tengcongensis Pep: Sugar Phosphotransferase System (Pts)
J.Mol.Biol., 346, 2005
|
|
2BGL
 
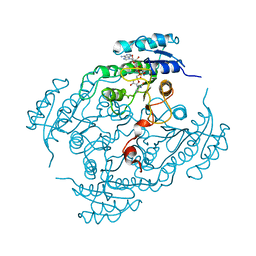 | | X-Ray structure of binary-Secoisolariciresinol Dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
2C3P
 
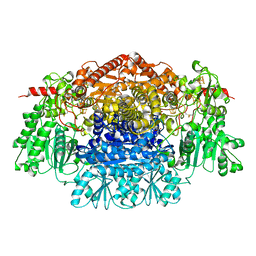 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 1-(2-{(2S,4R,5R)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-[(1S)-1-CARBOXY-1-HYDROXYETHYL]-4-METHYL-1,3-THIAZOLIDIN-5-YL}ETHOXY)-1,1,3,3-TETRAHYDROXY-1LAMBDA~5~-DIPHOSPHOX-1-EN-2-IUM 3-OXIDE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2BHJ
 
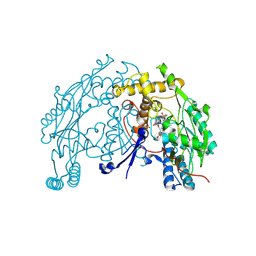 | | murine iNO synthase with coumarin inhibitor | | Descriptor: | 7,8-DIHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mathieu, M, Guilloteau, J.P. | | Deposit date: | 2005-01-12 | | Release date: | 2005-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design, Synthesis and Characterization of a Novel Class of Coumarin-Based Inhibitors of Inducible Nitric Oxide Synthase
Bioorg.Med.Chem., 13, 2005
|
|
2BI4
 
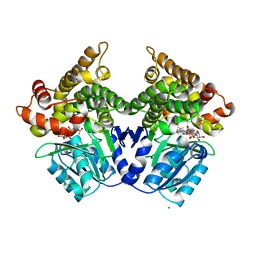 | | Lactaldehyde:1,2-propanediol oxidoreductase of Escherichia coli | | Descriptor: | CHLORIDE ION, FE (III) ION, LACTALDEHYDE REDUCTASE, ... | | Authors: | Montella, C, Bellsolell, L, Badia, J, Baldoma, L, Perez, R, Coll, M, Aguilar, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of an Iron-Dependent Group III Dehydrogenase that Interconverts L-Lactaldehyde and L-1,2-Propanediol in Escherichia Coli
J.Bacteriol., 187, 2005
|
|
6D3N
 
 | | Crystal structure of h4-1BB ligand | | Descriptor: | GLYCEROL, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
7A2F
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-656 | | Descriptor: | 1-cycloheptyl-3-{4-methoxy-3-[2-(4-methoxyphenyl)ethoxy]phenyl}-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-08-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-656
To be published
|
|
2BZ4
 
 | | structure of E.coli KAS I H298Q mutant | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE I, AMMONIUM ION, SULFATE ION | | Authors: | Olsen, J.G, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2005-08-10 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type beta-ketoacyl-acyl carrier protein synthases.
FEBS J., 273, 2006
|
|
2C5E
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), k217a, with gdp-alpha-d-mannose bound in the active site. | | Descriptor: | FORMIC ACID, GDP-MANNOSE-3', 5'-EPIMERASE, ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
6CB9
 
 | | Segment AALQSS from the low complexity domain of TDP-43, residues 328-333 | | Descriptor: | AALQSS | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2BJG
 
 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CHOLOYLGLYCINE HYDROLASE | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2BJV
 
 | | Crystal Structure of PspF(1-275) R168A mutant | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
6CN7
 
 | |
2BKJ
 
 | |
2BKQ
 
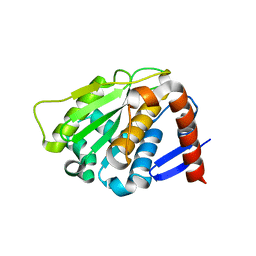 | | NEDD8 protease | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 8 | | Authors: | Shen, L.N, Liu, H, Dong, C, Xirodimas, D, Naismith, J.H, Hay, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Nedd8 Ubiquitin Discrimination by the Deneddylating Enzyme Nedp1
Embo J., 24, 2005
|
|
2C6K
 
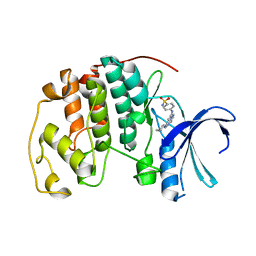 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLAMINO)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6CO6
 
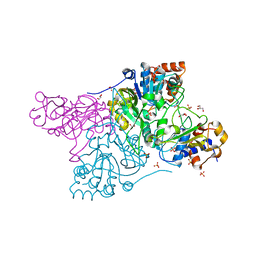 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BLH
 
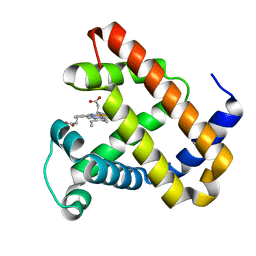 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
7A28
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-617 | | Descriptor: | 1-cycloheptyl-3-(4-methoxy-3-{4-[4-(1H-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy}phenyl)-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-617
To be published
|
|
2C1H
 
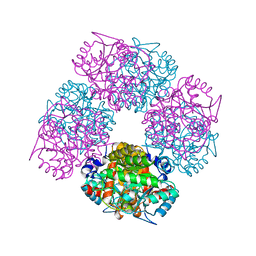 | | The X-ray Structure of Chlorobium vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor | | Descriptor: | 4,7-DIOXOSEBACIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S, Wood, S.P, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2005-09-14 | | Release date: | 2005-12-02 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Chlorobium Vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6CN5
 
 | |
2BL9
 
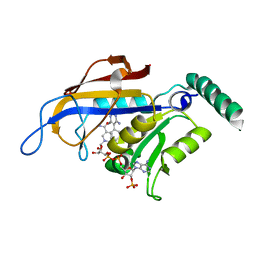 | | X-ray crystal structure of Plasmodium vivax dihydrofolate reductase in complex with pyrimethamine and its derivative | | Descriptor: | 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-02 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLQ
 
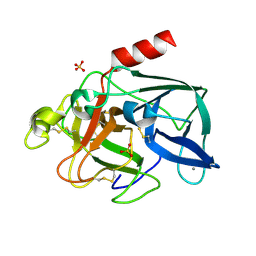 | | Elastase After A High Dose X-Ray "Burn" | | Descriptor: | CALCIUM ION, ELASTASE 1, SULFATE ION | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BNA
 
 | |
2BL1
 
 | | Crystal structure of a putative phosphinothricin Acetyltransferase (PA4866) from Pseudomonas aeruginosa PAC1 | | Descriptor: | AZIDE ION, GLYCEROL, PUTATIVE PHOSPHINOTHRICIN N-ACETYLTRANSFERASE PA4866, ... | | Authors: | Davies, A.M, Tata, R, Agha, R, Sutton, B.J, Brown, P.R. | | Deposit date: | 2005-02-24 | | Release date: | 2005-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Putative Phosphinothricin Acetyltransferase (Pa4866) from Pseudomonas Aeruginosa Pac1
Proteins: Struct., Funct., Bioinf., 61, 2005
|
|
