5FM9
 
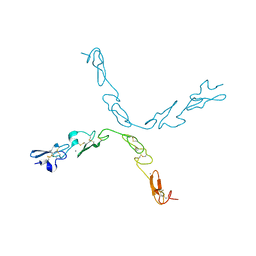 | | human Notch 1, EGF 4-7 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Weisshuhn, P.C, Sheppard, D, Taylor, P, Whiteman, P, Lea, S.M, Handford, P.A, Redfield, C. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Structure, 24, 2016
|
|
5OT9
 
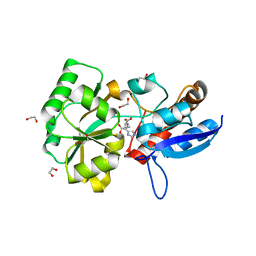 | |
6DFJ
 
 | |
5OTO
 
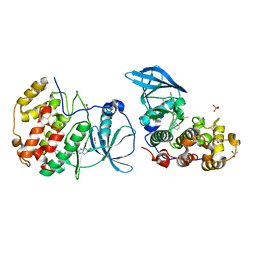 | | The crystal structure of CK2alpha in complex with compound 30 | | Descriptor: | 2-(5-chloranyl-1~{H}-benzimidazol-2-yl)-~{N}-[[3-chloranyl-4-(2-ethylphenyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
5SIY
 
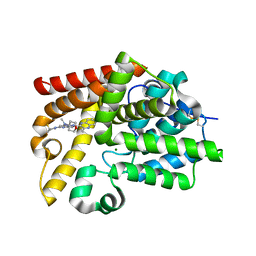 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nc2cc(nn12)c3c(nc4c(n3)cccc4)C)N5C[C@@H](CC5)F)N(C6CCOCC6)C, micromolar IC50=0.0050385 | | Descriptor: | (8R)-5-[(3R)-3-fluoropyrrolidin-1-yl]-N-methyl-2-(3-methylquinoxalin-2-yl)-N-(oxan-4-yl)pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Gobbi, L, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7AMC
 
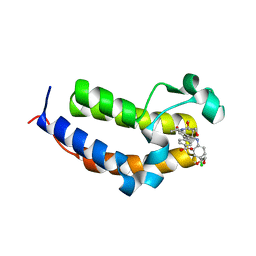 | | SmBRD3(2), Bromodomain 2 of the Bromodomain 3 protein from Schistosoma mansoni in complex with iBET726 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2S,4R)-1-acetyl-4-[(4-chlorophenyl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl]benzoic acid, GLYCEROL, ... | | Authors: | Schiedel, M, McDonough, M.A, Conway, S.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | SBM3, Bromodomain 3 from Schistosoma mansoni in complex with iBET726
To Be Published
|
|
6JP6
 
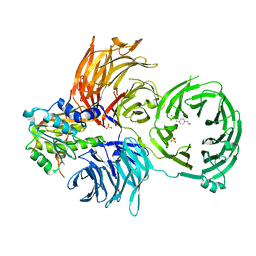 | | The X-ray structure of yeast tRNA methyltransferase complex of Trm7 and Trm734 essential for 2'-O-methylation at the first position of anticodon in specific tRNAs | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, tRNA (cytidine(34)/guanosine(34)-2'-O)-methyltransferase, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Sihimzu, N, Hori, H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
2VX4
 
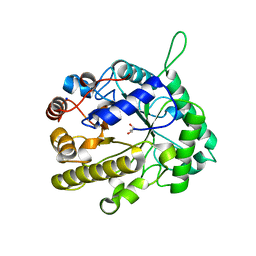 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C NATIVE FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, GLYCEROL, SODIUM ION | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
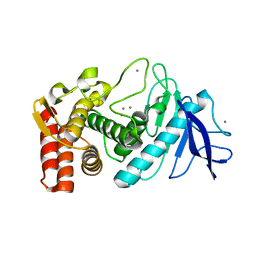
3P7T
 
 | |
4NV7
 
 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase 1 In Complex With CoA | | Descriptor: | Arylamine N-acetyltransferase, COENZYME A | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FW5
 
 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
6PUM
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-2'D-5-OP-RU | | Descriptor: | 1,2-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
5SIU
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2c(Cl)c1nc(nn1c(c2Cl)C)CCC3=Nc4c(NC3=O)cccc4, micromolar IC50=0.007718 | | Descriptor: | 3-{2-[(4S)-6,8-dichloro-5-methyl[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethyl}quinoxalin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7AQL
 
 | |
5SIS
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(n(nc1)C)C(Nc3cc2nc(cn2cc3)c4ccccc4)=O)C(=O)N(C)CCNC, micromolar IC50=0.002096 | | Descriptor: | MAGNESIUM ION, N~4~,1-dimethyl-N~4~-[2-(methylamino)ethyl]-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7X8K
 
 | | Arabidopsis GDP-D-mannose pyrophosphorylase (VTC1) structure (product-bound) | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, Mannose-1-phosphate guanylyltransferase 1, ... | | Authors: | Zhao, S, Zhang, C, Liu, L. | | Deposit date: | 2022-03-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana GDP-D-Mannose Pyrophosphorylase VITAMIN C DEFECTIVE 1.
Front Plant Sci, 13, 2022
|
|
4IBS
 
 | | Human p53 core domain with hot spot mutation R273H (form I) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
5SK7
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(cc1nc(nn1cc2)N(C)C)Br, micromolar IC50=4.393076 | | Descriptor: | 4-bromanyl-~{N},~{N}-dimethyl-1$l^{4},7,9-triazabicyclo[4.3.0]nona-1,3,5-trien-8-amine, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4NZY
 
 | | Crystal structure (type-2) of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into Substrate Binding Mechanism in Thymidylate Kinase based on Intrinsic Dynamics of the Active Site
To be Published
|
|
5SJU
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(nc(s1)NC(c2c(ncc(n2)C3CC3)Nc4cncnc4)=O)CN5CCCCC5, micromolar IC50=0.03208 | | Descriptor: | 6-cyclopropyl-N-{4-[(piperidin-1-yl)methyl]-1,3-thiazol-2-yl}-3-[(pyrimidin-5-yl)amino]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7X8J
 
 | |
1UGU
 
 | | Crystal structure of PYP E46Q mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Sugishima, M, Tanimoto, Y, Hamada, N, Tokunaga, F, Fukuyama, K. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MM7
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Quisqualate in a Zinc Crystal Form at 1.65 Angstroms Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Activation and Selectivity in a Ligand-Gated Ion Channel: Structural and Functional Studies of GluR2 and Quisqualate
Biochemistry, 41, 2003
|
|
7I1W
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with 3632-JP-070-010 | | Descriptor: | (1r,4r)-4-amino-N-[4-(hydroxymethyl)-2-methylquinolin-8-yl]cyclohexane-1-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2WK0
 
 | | Crystal structure of the class A beta-lactamase BS3 inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Sauvage, E, Zervosen, A, Dive, G, Herman, R, Kerff, F, Amoroso, A, Fonze, E, Pratt, R.F, Luxen, A, Charlier, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
