7P72
 
 | | The PDZ domain of SNX27 complexed with the PDZ-binding motif of MERS-E | | Descriptor: | CALCIUM ION, Envelope small membrane protein, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P73
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, GLYCEROL, Protein Tax-1, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P74
 
 | | The PDZ domain of SYNJ2BP complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Ribosomal protein S6 kinase alpha-1, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P70
 
 | | The PDZ-domain of SNTB1 complexed with the PDZ-binding motif of HPV35-E6 | | Descriptor: | Beta-1-syntrophin,Annexin A2, CALCIUM ION, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P71
 
 | | The PDZ domain of MAGI1_2 complexed with the PDZ-binding motif of HPV35-E6 | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
5YDF
 
 | | Crystal structure of a disease-related gene, hCDC73(1-100) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
6VZ4
 
 | |
5YBD
 
 | | X-ray structure of ETS domain of Ergp55 in complex with E74DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8USC
 
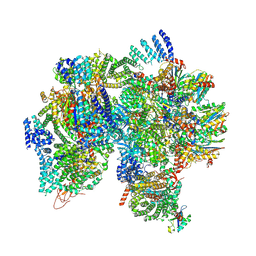 | | Nub1/Fat10-processing human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Arkinson, C, Gee, C.L, Martin, A. | | Deposit date: | 2023-10-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | NUB1 traps unfolded FAT10 for ubiquitin-independent degradation by the 26S proteasome.
Nat.Struct.Mol.Biol., 2025
|
|
5L4G
 
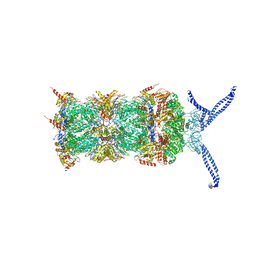 | | The human 26S proteasome at 3.9 A | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Aufderheide, A, Rudack, T, Beck, F. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human 26S proteasome at a resolution of 3.9 angstrom.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7VVZ
 
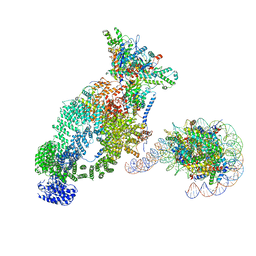 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
1DU0
 
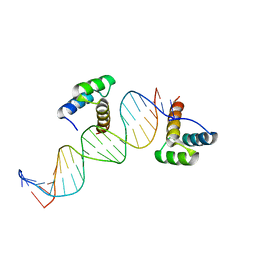 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1DY8
 
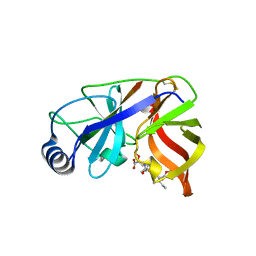 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
8H1T
 
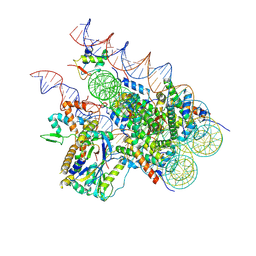 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | Descriptor: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | Authors: | Ge, W, Yu, C, Xu, R.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
8VWV
 
 | | OGG1 bound to a nucleosome containing 8oxoG at SHL4 (composite map) | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWT
 
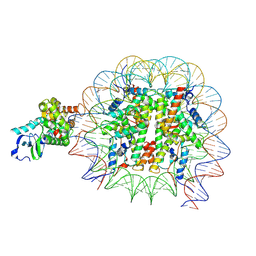 | | OGG1 bound to a nucleosome containing 8oxoG at SHL-6 (composite map) | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8RHZ
 
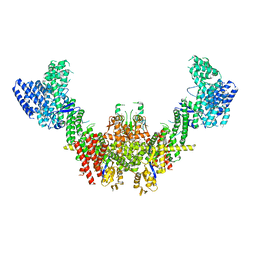 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7PC8
 
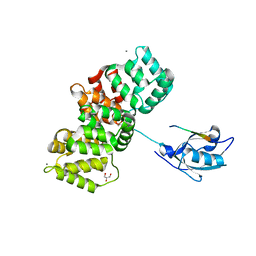 | | The PDZ domain of SNTG1 complexed with the phosphomimetic mutant PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PCB
 
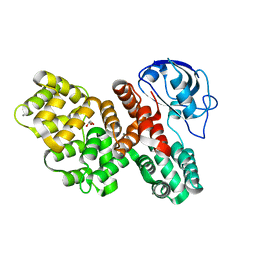 | | The PDZ domain of SNX27 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Sorting nexin-27,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC7
 
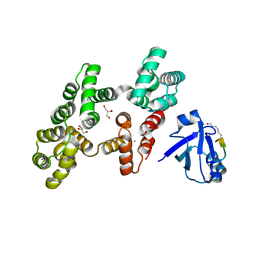 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of PTEN | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC9
 
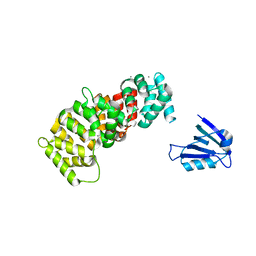 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Protein Tax-1, Synaptojanin-2-binding protein,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC3
 
 | | The second PDZ domain of DLG1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Disks large homolog 1,Annexin A2, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQL
 
 | | The PDZ domain of SNTG2 complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-2-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQM
 
 | | The PDZ domain of LRRC7 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Leucine-rich repeat-containing protein 7,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQN
 
 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of TRPV3 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
