1KKO
 
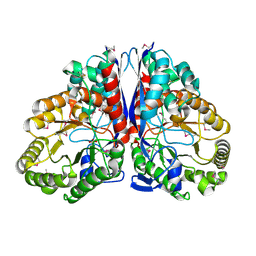 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KLJ
 
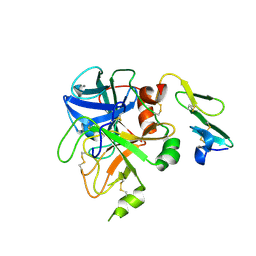 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
1KL2
 
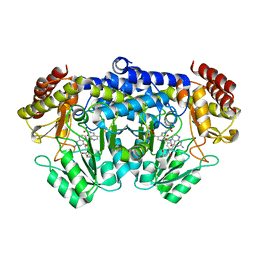 | | Crystal Structure of Serine Hydroxymethyltransferase Complexed with Glycine and 5-formyl tetrahydrofolate | | Descriptor: | GLYCINE, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-11 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KIR
 
 | |
1KV5
 
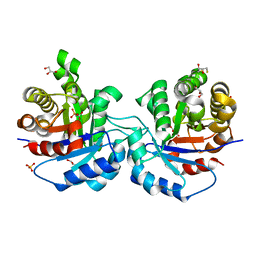 | | Structure of Trypanosoma brucei brucei TIM with the salt-bridge-forming residue Arg191 mutated to Ser | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ... | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The importance of the conserved Arg191-Asp227 salt bridge of triosephosphate isomerase for folding, stability, and catalysis
FEBS Lett., 518, 2002
|
|
2Y4D
 
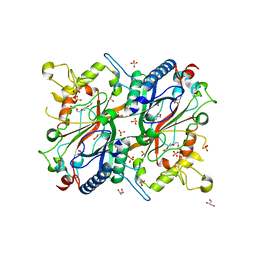 | | X-ray crystallographic structure of E. coli apo-EfeB | | Descriptor: | ACETATE ION, GLYCEROL, PEROXIDASE YCDB, ... | | Authors: | Bamford, V.A, Andrews, S.C, Watson, K.A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Efeb, the Peroxidase Component of the Efeuob Bacterial Fe(II) Transport System, Also Shows Novel Removal of Iron from Heme
To be Published
|
|
1KVB
 
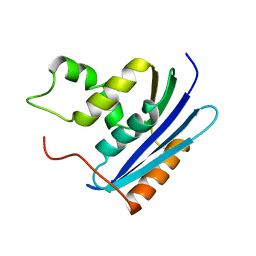 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
7AOK
 
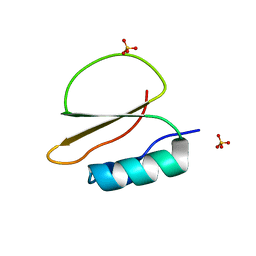 | | Crystal structure of CI2 mutant L49I | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
1KVM
 
 | | X-ray Crystal Structure of AmpC WT beta-Lactamase in Complex with Covalently Bound Cephalothin | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
7AP3
 
 | | Crystal structure of E. coli tyrosyl-tRNA synthetase in complex with TyrS7HMDDA | | Descriptor: | SODIUM ION, Tyrosine--tRNA ligase, [(2~{R},3~{S},4~{R},5~{R})-5-[7-azanyl-5-(hydroxymethyl)benzimidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(2~{S})-2-azanyl-3-(4-hydroxyphenyl)propanoyl]sulfamate | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
1KVY
 
 | |
1KLY
 
 | |
1KM4
 
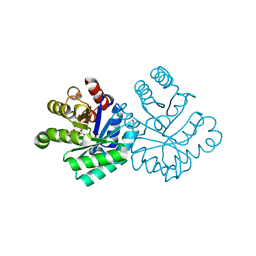 | |
7AQ0
 
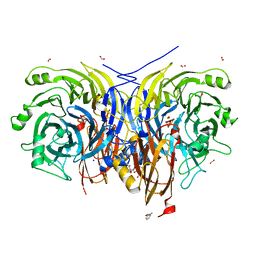 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A/S550A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1KLS
 
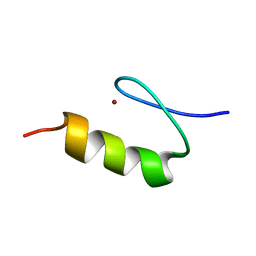 | | NMR Structure of the ZFY-6T[Y10L] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1KMC
 
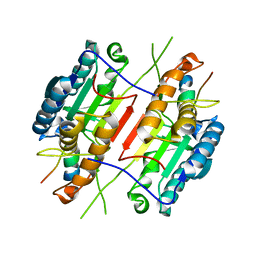 | |
9COO
 
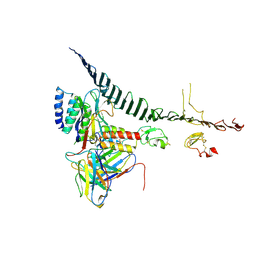 | | Nanobody 4 bound to Apolipoprotein B 100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, Apolipoprotein B 100, ... | | Authors: | Dearborn, A.D, Kumar, A, Reimund, M, Graziano, G, Lei, H, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-07-17 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
1KMH
 
 | |
1KMV
 
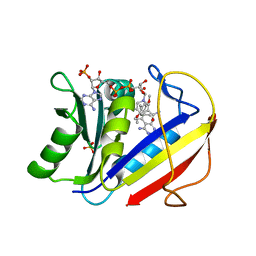 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9662), A LIPOPHILIC ANTIFOLATE | | Descriptor: | (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, DIMETHYL SULFOXIDE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
1KLA
 
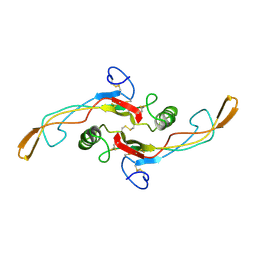 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KWE
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CONSERVED REGION OF HUMAN RESPIRATORY SYNCYTIAL VIRUS ATTACHMENT GLYCOPROTEIN G | | Descriptor: | MAJOR SURFACE GLYCOPROTEIN G | | Authors: | Sugawara, M, Czaplicki, J, Ferrage, J, Haeuw, J.F, Power, U.F, Corvaia, N, Nguyen, T, Beck, A, Milon, A. | | Deposit date: | 2002-01-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G.
J.Pept.Res., 60, 2002
|
|
2Y4R
 
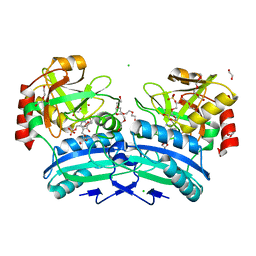 | | CRYSTAL STRUCTURE OF 4-AMINO-4-DEOXYCHORISMATE LYASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-4-DEOXYCHORISMATE LYASE, CHLORIDE ION, ... | | Authors: | O'Rourke, P.E.F, Eadsforth, T.C, Fyfe, P.K, Shepard, S.M, Agacan, M, Hunter, W.N. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pseudomonas Aeruginosa 4-Amino-4-Deoxychorismate Lyase: Spatial Conservation of an Active Site Tyrosine and Classification of Two Types of Enzyme.
Plos One, 6, 2011
|
|
1KWJ
 
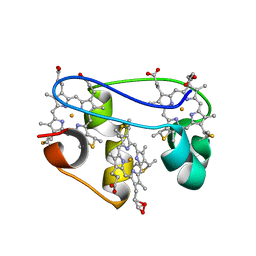 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1KWT
 
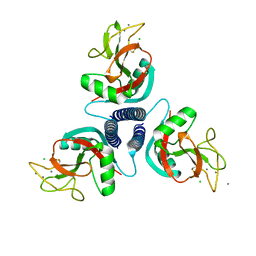 | | Rat mannose binding protein A (native, MPD) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A | | Authors: | Ng, K.K.S, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KMZ
 
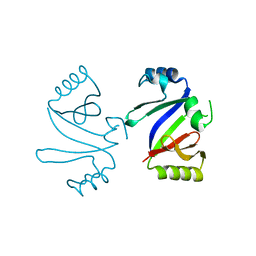 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
