1KR8
 
 | |
6W8W
 
 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
1KRL
 
 | | Crystal Structure of Racemic DL-monellin in P-1 | | Descriptor: | MONELLIN, CHAIN A, CHAIN B | | Authors: | Hung, L.W, Kohmura, M, Ariyoshi, Y, Kim, S.H. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural differences in D and L-monellin in the crystals of racemic mixture.
J.Mol.Biol., 285, 1999
|
|
1KJG
 
 | |
7AYH
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
6W2M
 
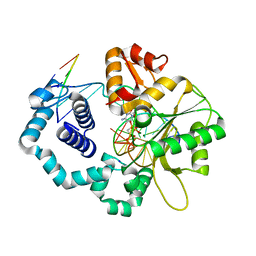 | |
1KJP
 
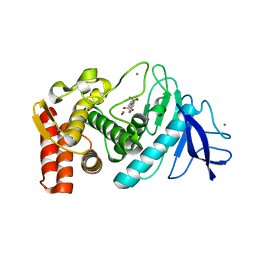 | |
1KRO
 
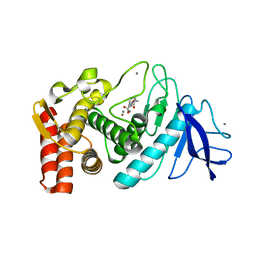 | |
1KSL
 
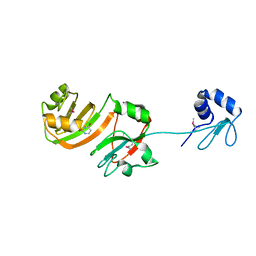 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KJI
 
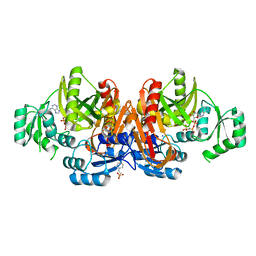 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
7B2S
 
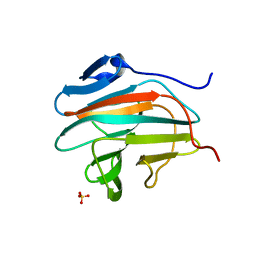 | |
1KJX
 
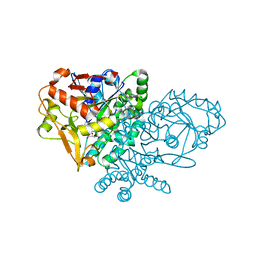 | | IMP Complex of E. Coli Adenylosuccinate Synthetase | | Descriptor: | Adenylosuccinate Synthetase, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KK6
 
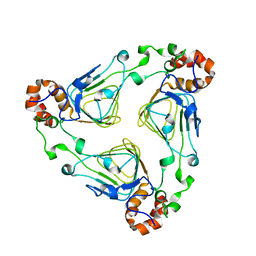 | | Crystal Structure of Vat(D) (Form I) | | Descriptor: | STREPTOGRAMIN A ACETYLTRANSFERASE | | Authors: | Sugantino, M, Roderick, S.L. | | Deposit date: | 2001-12-06 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics.
Biochemistry, 41, 2002
|
|
1KSV
 
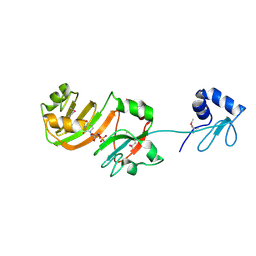 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KT0
 
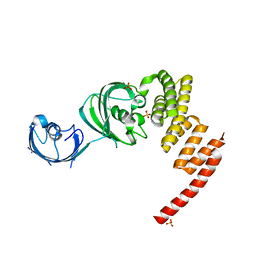 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | 51 KDA FK506-BINDING PROTEIN, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a
component of steroid receptor complexes
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7AKN
 
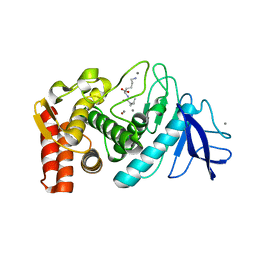 | | Thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Boikova, A.S, Ilina, K.B, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2020-10-01 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Thermolysin from Bacillus thermoproteolyticus
To Be Published
|
|
7AKW
 
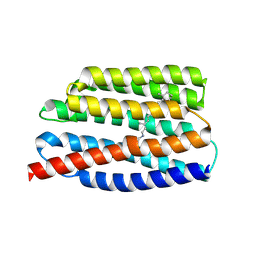 | | Crystal structure of the viral rhodopsins chimera O1O2 | | Descriptor: | EICOSANE, RETINAL, chimera of viral rhodopsins OLPVR1 and OLPVRII | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
1KTQ
 
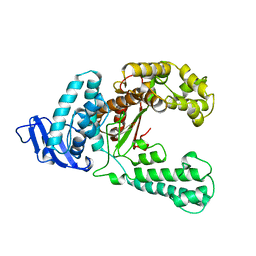 | | DNA POLYMERASE | | Descriptor: | DNA POLYMERASE I | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1995-08-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7AKY
 
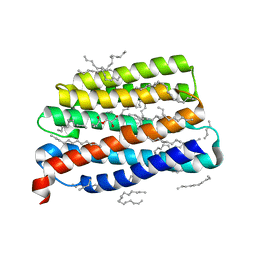 | | Crystal structure of the viral rhodopsin OLPVR1 in P21212 space group | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, viral rhodopsin OLPVR1 | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
1KK7
 
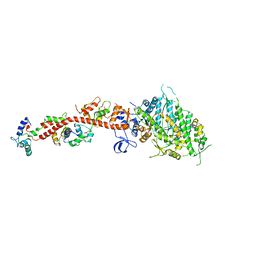 | | SCALLOP MYOSIN IN THE NEAR RIGOR CONFORMATION | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MYOSIN ESSENTIAL LIGHT CHAIN, ... | | Authors: | Himmel, D.M, Gourinath, S, Reshetnikova, L, Shen, Y, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2001-12-06 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7B3Z
 
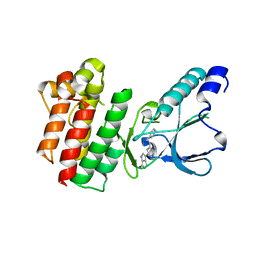 | | Crystal structure of c-MET bound by compound 5 | | Descriptor: | 3-[3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
1KKL
 
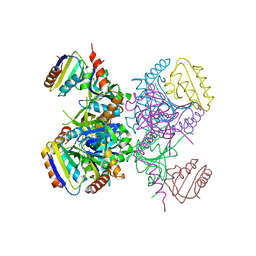 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KKW
 
 | |
1KU9
 
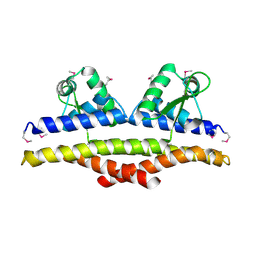 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
1KUX
 
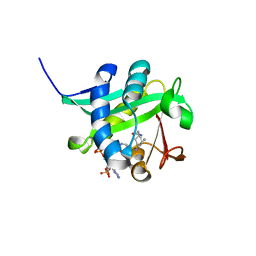 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-TRIMETHYLENE-ACETYL-TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
