1J4Q
 
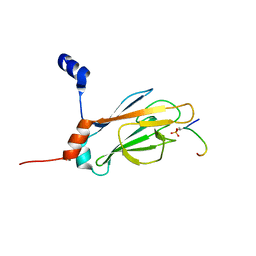 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1JQR
 
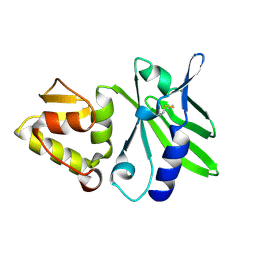 | |
1NYG
 
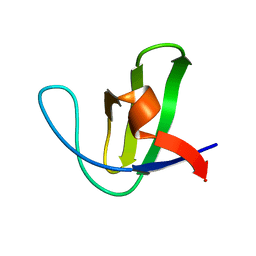 | |
2OJ2
 
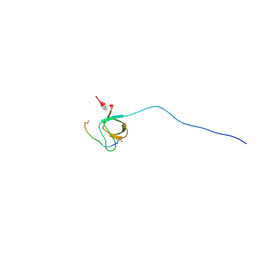 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Hematopoetic Cell Kinase, SH3 domain, artificial peptide PD1 | | Authors: | Schmidt, H, Hoffmann, S, Tran, T, Stoldt, M, Stangler, T, Wiesehan, K, Willbold, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
1NYF
 
 | |
1J4P
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1UAO
 
 | |
1TR6
 
 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1KKV
 
 | |
1KKW
 
 | |
1IN1
 
 | |
2LQP
 
 | |
2LWZ
 
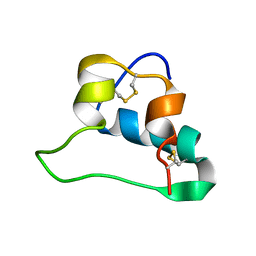 | | NMR Structures of Single-chain Insulin | | Descriptor: | SINGLE-CHAIN INSULIN | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2012-08-09 | | Release date: | 2013-08-28 | | Method: | SOLUTION NMR | | Cite: | Dynamic repair of an amyloidogenic protein: Insulin fibrillation is blocked by tethering a nascent alpha-helix
To be Published
|
|
2N17
 
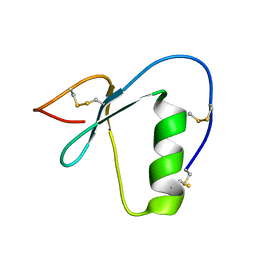 | | NMR structure of a Kazal-type serine protease inhibitor from the subterranean termite defense gland of Coptotermes formosanus Shiraki soldiers | | Descriptor: | Lysozyme-Protease Inhibitor Protein | | Authors: | Negulescu, H, Guo, Y, Garner, T.P, Goodwin, O.Y, Henderson, G, Laine, R.A, Macnaughtan, M.A. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Kazal-Type Serine Protease Inhibitor from the Defense Gland Secretion of the Subterranean Termite Coptotermes formosanus Shiraki.
Plos One, 10, 2015
|
|
2N7L
 
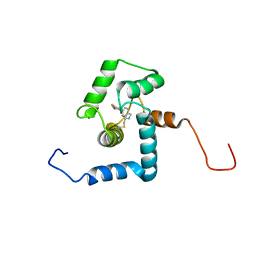 | |
2N01
 
 | | NMR structure of VirB9 C-terminal domain in complex with VirB7 N-terminal domain from Xanthomonas citri's T4SS | | Descriptor: | VirB7 protein, VirB9 protein | | Authors: | Oliveira, L.C, Souza, D.P, Salinas, R.K, Wienk, H, Boelens, R, Farah, S.C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | VirB7 and VirB9 Interactions Are Required for the Assembly and Antibacterial Activity of a Type IV Secretion System.
Structure, 24, 2016
|
|
2OI3
 
 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Tyrosine-protein kinase HCK, artificial peptide PD1 | | Authors: | Schmidt, H, Stoldt, M, Hoffmann, S, Tran, T, Willbold, D. | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand
Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
2NO8
 
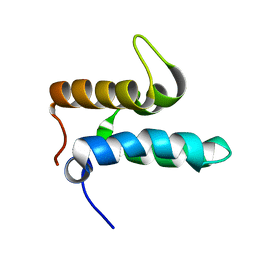 | |
1DSJ
 
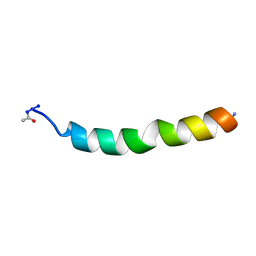 | | NMR SOLUTION STRUCTURE OF VPR50_75, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Helical structure of polypeptides from the C-terminal half of HIV-1 VPR.
Protein Pept.Lett., 5, 1998
|
|
1EVC
 
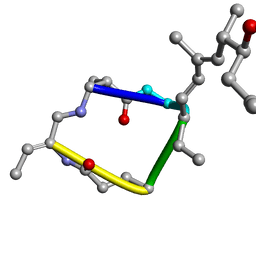 | |
1EVB
 
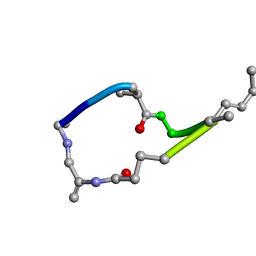 | |
1EMN
 
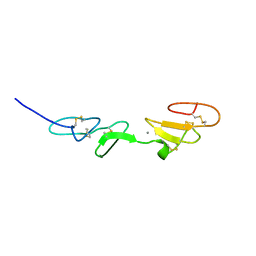 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
1EMO
 
 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, 22 STRUCTURES | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
1FOX
 
 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
