6MYY
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
4Q53
 
 | | Crystal structure of a DUF4783 family protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of a hypothetical protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution
To be published
|
|
4Q7V
 
 | |
4Q83
 
 | |
4Q8X
 
 | |
3LM2
 
 | |
3TKW
 
 | | Crystal structure of HIV protease model precursor/Darunavir complex | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
5F1Z
 
 | |
1H0W
 
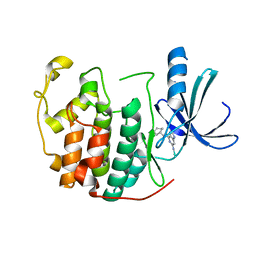 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[cyclohex-3-enyl]methoxypurine | | Descriptor: | 1-AMINO-6-CYCLOHEX-3-ENYLMETHYLOXYPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
6MN7
 
 | | Cryo-EM structure of BG505.SOSIP.664 in complex with BF520.1 antigen binding fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BF520.1 Fab variable region, ... | | Authors: | Williams, J.A, Lee, K.K, Overbaugh, J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage.
Nat Commun, 10, 2019
|
|
5FXQ
 
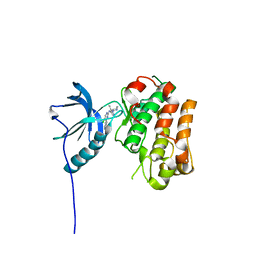 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 5-chloranyl-4-imidazo[1,2-a]pyridin-3-yl-N-(5-methyl-1-piperidin-4-yl-pyrazol-4-yl)pyrimidin-2-amine, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
1GZ8
 
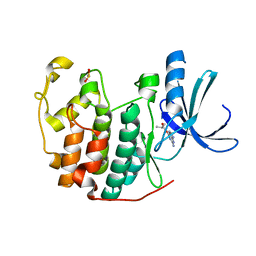 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 2-Amino-6-(3'-methyl-2'-oxo)butoxypurine | | Descriptor: | 1-[(2-AMINO-6,9-DIHYDRO-1H-PURIN-6-YL)OXY]-3-METHYL-2-BUTANOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davies, T, Endicott, J, Johnson, L, Noble, M, Tucker, J. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
3ZYQ
 
 | | Crystal structure of the tandem VHS and FYVE domains of Hepatocyte growth factor-regulated tyrosine kinase substrate (HGS-Hrs) at 1.48 A resolution | | Descriptor: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Ayinampudi, V, Shrestha, L, Krojer, T, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) at 1.48 A Resolution
To be Published
|
|
5CR5
 
 | |
6DUC
 
 | | Crystal structure of mutant beta-K167T tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid (1D0) at the beta-site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate (P1T) at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
4YDF
 
 | | Crystal structure of compound 9 in complex with HTLV-1 Protease | | Descriptor: | HTLV-1 Protease, N-benzyl-N-[(3S,4S)-4-{benzyl[(4-nitrophenyl)sulfonyl]amino}pyrrolidin-3-yl]-3-nitrobenzenesulfonamide, SULFATE ION | | Authors: | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | Deposit date: | 2015-02-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YTR
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
3TL9
 
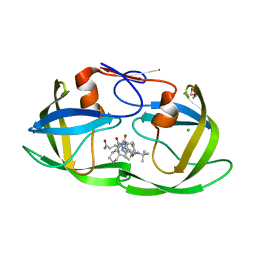 | | crystal structure of HIV protease model precursor/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
6T5K
 
 | |
5GSS
 
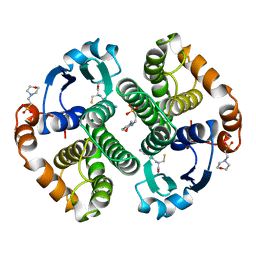 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
5D3T
 
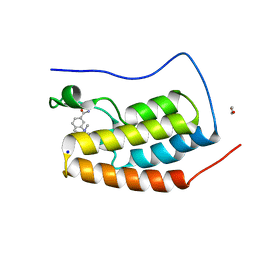 | | First bromodomain of BRD4 bound to inhibitor XD47 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-(3-carbamoylbenzyl)-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
4YTQ
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy D-tagatose | | Descriptor: | 1-deoxy-D-tagatose, 1-deoxy-alpha-D-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5ZT0
 
 | |
2WIY
 
 | | Cytochrome P450 XplA heme domain P21212 | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450-LIKE PROTEIN XPLA, IMIDAZOLE, ... | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
3IIC
 
 | |
