7JJ6
 
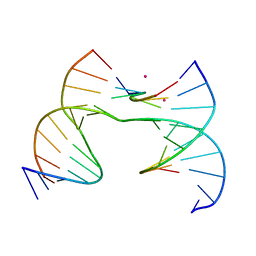 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J8 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.077 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JK0
 
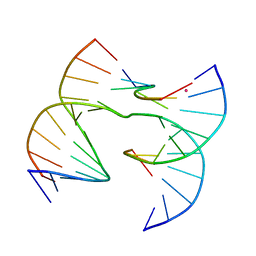 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*TP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.058 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JIO
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J23 immobile Holliday junction with R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*GP*TP*GP*TP*CP*GP*T)-3'), DNA (5'-D(P*CP*GP*AP*CP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJ5
 
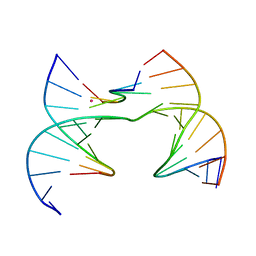 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J10 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJX
 
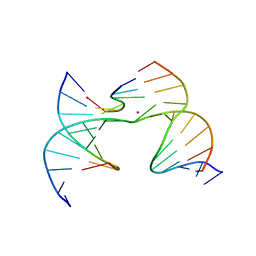 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J5 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFT
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J2 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JH9
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J22 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHR
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J4 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJ4
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J14 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*CP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.022 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
3OOB
 
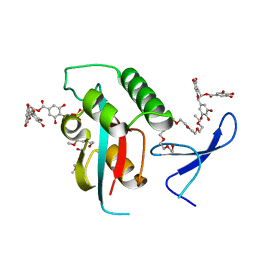 | | Structural and functional insights of directly targeting Pin1 by Epigallocatechin-3-gallate | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Urusova, D.V, Shim, J.-H, Kim, D.-J, Jung, S.K, Zykova, T.A, Bode, A.M, Dong, Z. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1.
Cancer Prev Res (Phila), 4, 2011
|
|
169D
 
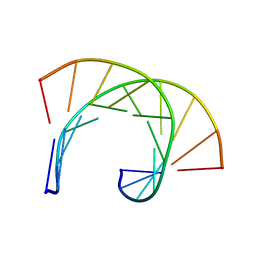 | | THE SOLUTION STRUCTURE OF THE R(GCG)D(TATACCC):D(GGGTATACGC) OKAZAKI FRAGMENT CONTAINS TWO DISTINCT DUPLEX MORPHOLOGIES CONNECTED BY A JUNCTION | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*TP*AP*CP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*G)-D(P*TP*AP*TP*AP*CP*CP*C)-3') | | Authors: | Salazar, M, Fedoroff, O.Y, Zhu, L, Reid, B.R. | | Deposit date: | 1994-04-11 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction.
J.Mol.Biol., 241, 1994
|
|
7JKD
 
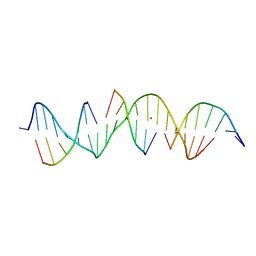 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*GP*AP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.058 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKE
 
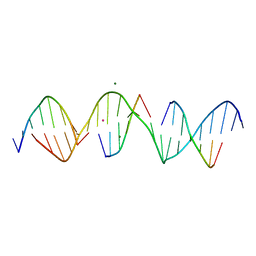 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J2 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKI
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J7 immobile Holliday junction with R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*AP*GP*AP*CP*GP*GP*TP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKG
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J3 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*GP*TP*GP*AP*CP*GP*TP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKK
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J10 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*TP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKH
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J5 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*CP*GP*AP*CP*GP*GP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKJ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J8 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*AP*GP*AP*CP*GP*AP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.077 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
1M6G
 
 | | Structural Characterisation of the Holliday Junction TCGGTACCGA | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', STRONTIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-07-16 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Conformational and hydration effects of site-selective sodium, calcium and
strontium ion binding to the DNA Holliday junction structure
d(TCGGTACCGA)(4)
J.Mol.Biol., 327, 2003
|
|
2LA5
 
 | | RNA Duplex-Quadruplex Junction Complex with FMRP RGG peptide | | Descriptor: | Fragile X mental retardation 1 protein, RNA (36-MER) | | Authors: | Phan, A, Kuryavyi, V, Darnell, J, Serganov, A, Majumdar, A, Ilin, S, Darnell, R, Patel, D. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1KBU
 
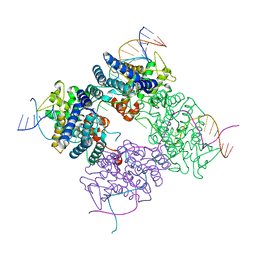 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
1L4J
 
 | | Holliday Junction TCGGTACCGA with Na and Ca Binding Sites. | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', CALCIUM ION, SODIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-03-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational and Hydration Effects of Site-selective Sodium, Calcium and
Strontium Ion Binding to the DNA Holliday Junction Structure
d(TCGGTACCGA)4
J.Mol.Biol., 327, 2003
|
|
196D
 
 | |
1IN7
 
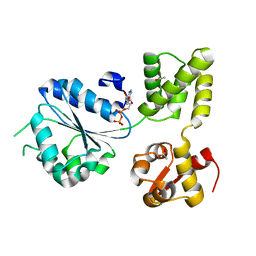 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
