2QW5
 
 | |
2R3S
 
 | |
2QYC
 
 | |
3H7C
 
 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2R44
 
 | |
2QR6
 
 | |
2QVP
 
 | |
2QWW
 
 | |
2R3B
 
 | |
2QCV
 
 | |
2QE6
 
 | |
2QEZ
 
 | |
2QJ8
 
 | |
2QTQ
 
 | |
2QYV
 
 | |
2R1I
 
 | |
2QCK
 
 | |
3POU
 
 | |
3PUM
 
 | | Crystal structure of P domain dimer of Norovirus VA207 | | Descriptor: | Capsid | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3QNS
 
 | | DyPB from Rhodococcus jostii RHA1, crystal form 2 | | Descriptor: | DyP Peroxidase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Grigg, J.C, Roberts, J.N, Singh, R, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1.
Biochemistry, 50, 2011
|
|
3QPT
 
 | |
3QMB
 
 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3', CALCIUM ION, CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3QNQ
 
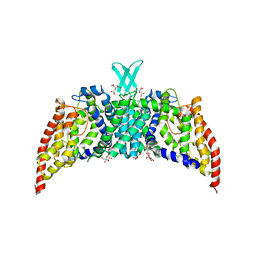 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
