2IF1
 
 | | HUMAN TRANSLATION INITIATION FACTOR EIF1, NMR, 29 STRUCTURES | | Descriptor: | EIF1 | | Authors: | Fletcher, C.M, Hellen, C.U.T, Pestova, T.V, Wagner, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the translation initiation factor eIF1.
EMBO J., 18, 1999
|
|
2JNC
 
 | | Refined 3D NMR structure of ECD1 of mCRF-R2beta at pH 5 | | Descriptor: | Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JND
 
 | | 3D NMR structure of ECD1 of mCRF-R2b in complex with Astressin | | Descriptor: | ASTRESSIN, Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2KN0
 
 | | Solution NMR Structure of xenopus Fn14 | | Descriptor: | Fn14 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-06-29 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2I19
 
 | | T. Brucei farnesyl diphosphate synthase complexed with bisphosphonate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [2-(PYRIDIN-2-YLAMINO)ETHANE-1,1-DIYL]BIS(PHOSPHONIC ACID) | | Authors: | Cao, R, Mao, J, Gao, Y, Robinson, H, Odeh, S, Goddard, A, Oldfield, E. | | Deposit date: | 2006-08-13 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Solid-state NMR, crystallographic, and computational investigation of bisphosphonates and farnesyl diphosphate synthase-bisphosphonate complexes.
J.Am.Chem.Soc., 128, 2006
|
|
2KTX
 
 | | COMPLETE KALIOTOXIN FROM ANDROCTONUS MAURETANICUS MAURETANICUS, NMR, 18 STRUCTURES | | Descriptor: | KALIOTOXIN | | Authors: | Gairi, M, Romi, R, Fernandez, I, Rochat, H, Martin-Eauclaire, M.-F, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | 3D structure of kaliotoxin: is residue 34 a key for channel selectivity?
J.Pept.Sci., 3, 1997
|
|
2FOW
 
 | | THE RNA BINDING DOMAIN OF RIBOSOMAL PROTEIN L11: THREE-DIMENSIONAL STRUCTURE OF THE RNA-BOUND FORM OF THE PROTEIN, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL PROTEIN L11 | | Authors: | Hinck, A.P, Markus, M.A, Huang, S, Grzesiek, S, Kustanovich, I, Draper, D.E, Torchia, D.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA.
J.Mol.Biol., 274, 1997
|
|
2K38
 
 | | Cupiennin 1A, NMR, minimized average structure | | Descriptor: | Cupiennin-1a | | Authors: | Pukala, T.L, Boland, M.P, Gehman, J.D, Kuhn-Nentwig, L, Separovic, F, Bowie, J.H. | | Deposit date: | 2008-04-23 | | Release date: | 2008-05-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction of cupiennin 1a, a spider venom peptide, with phospholipid bilayers
Biochemistry, 46, 2007
|
|
2L0W
 
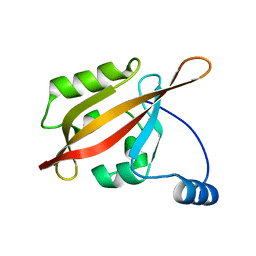 | | Solution NMR structure of the N-terminal PAS domain of HERG potassium channel | | Descriptor: | Potassium voltage-gated channel, subfamily H (Eag-related), member 2, ... | | Authors: | Ng, C.A, Hunter, M.J, Mobli, M, King, G.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Tail of hERG Contains an Amphipathic alpha-Helix That Regulates Channel Deactivation
PLoS ONE, 6, 2011
|
|
2EWA
 
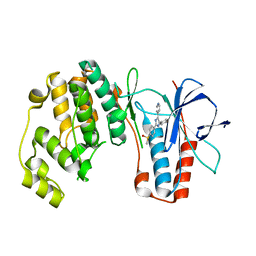 | | Dual binding mode of pyridinylimidazole to MAP kinase p38 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | Delarbre, L, Pouzieux, S, Guilloteau, J.-P, Michot, N. | | Deposit date: | 2005-11-02 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NMR characterization of kinase p38 dynamics in free and ligand-bound forms.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2IGG
 
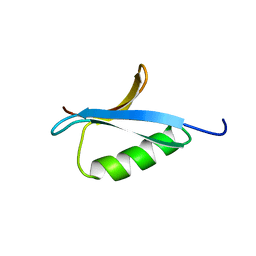 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
2K0T
 
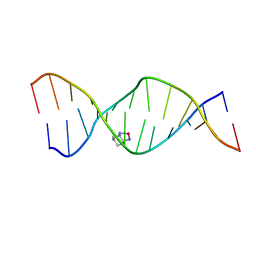 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-14 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2HYS
 
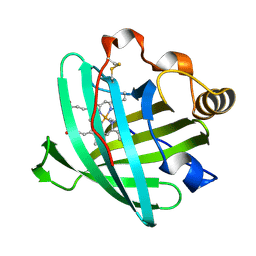 | | Crystal structure of nitrophorin 2 complexed with cyanide | | Descriptor: | CYANIDE ION, Nitrophorin-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Assignment of the Ferriheme Resonances of the Low-Spin Complexes of Nitrophorins 1 and 4 by (1)H and (13)C NMR Spectroscopy: Comparison to Structural Data Obtained from X-ray Crystallography.
Inorg.Chem., 46, 2007
|
|
2HCC
 
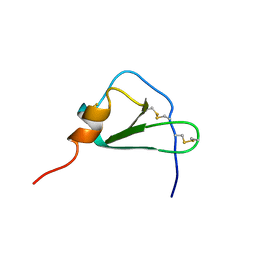 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | Descriptor: | HUMAN CHEMOKINE HCC-2 | | Authors: | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | Deposit date: | 1998-07-03 | | Release date: | 1999-07-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
2HFH
 
 | |
2KXO
 
 | |
2GCC
 
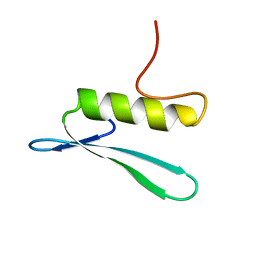 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
2KEF
 
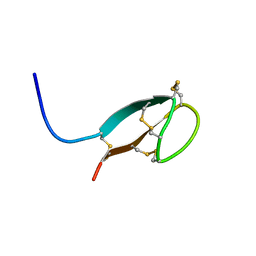 | | Solution NMR structures of human hepcidin at 325K | | Descriptor: | Hepcidin | | Authors: | Jordan, J.B, Poppe, L, Hainu, M, Arvedson, T, Syed, R, Li, V, Kohno, H, Kim, H, Miranda, L.P, Cheetham, J, Sasu, B.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2KMJ
 
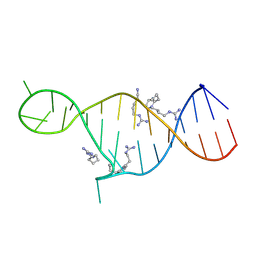 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
2H95
 
 | |
2LR9
 
 | |
2LIA
 
 | | Solution NMR structure of a DNA dodecamer containing the 7-aminomethyl-7-deaza-2'-deoxyguanosine adduct | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*(2LA)P*CP*GP*CP*TP*CP*TP*C)-3') | | Authors: | Szulik, M.W, Ganguly, M, Wang, R, Gold, B, Stone, M.P. | | Deposit date: | 2011-08-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Site-Specific Stabilization of DNA by a Tethered Major Groove Amine, 7-Aminomethyl-7-deaza-2'-deoxyguanosine.
Biochemistry, 52, 2013
|
|
2JSH
 
 | | obestatin NMR structure in SDS/DPC micellar solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2K0U
 
 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2K0V
 
 | | High Resolution Solution NMR Structures of Undamaged DNA Dodecamer Duplex | | Descriptor: | DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
