4XJ1
 
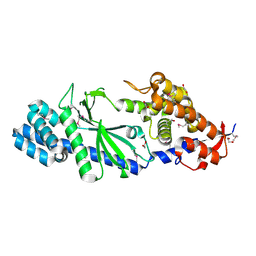 | | Crystal structure of Vibrio cholerae DncV apo form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
7RIX
 
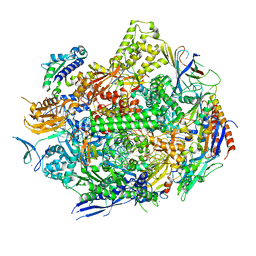 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5AOJ
 
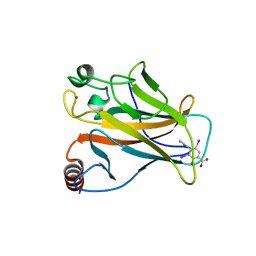 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
7ZQT
 
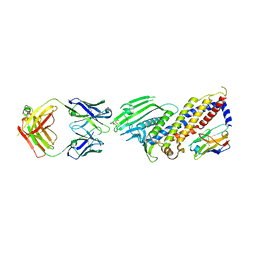 | |
6P2R
 
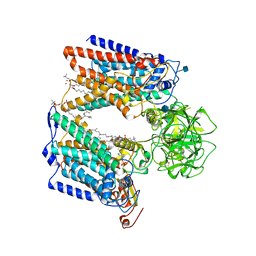 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3GUJ
 
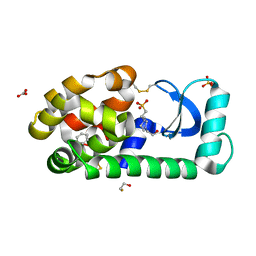 | |
4QBW
 
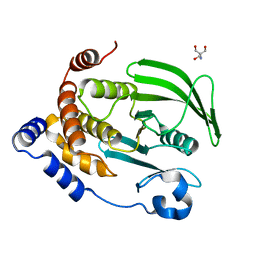 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X, Sun, J.P. | | Deposit date: | 2014-05-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57, 2014
|
|
9C3W
 
 | | Crystal structure of biphenyl synthase from Malus domestica complexed with diketide-CoA mimetics | | Descriptor: | (3R)-butane-1,3-diol, BIS3 biphenyl synthase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-[(3-oxo-3-{[2-(3-oxo-3-phenylpropanamido)ethyl]amino}propyl)amino]butyl dihydrogen diphosphate, ... | | Authors: | Re, R.N, Noel, J.P, Burkart, M.D. | | Deposit date: | 2024-06-02 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Elucidating the Iterative Elongation Mechanism in a Type III Polyketide Synthase.
J.Am.Chem.Soc., 147, 2025
|
|
6RFZ
 
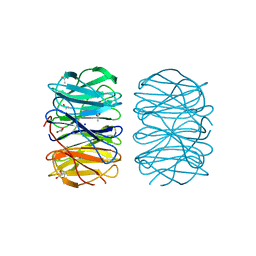 | | Photorhabdus laumondii lectin PLL2 in complex with D-glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SODIUM ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
7RIM
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6RG1
 
 | | Photorhabdus laumondii lectin PLL2 in complex with L-rhamnose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SODIUM ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
5F1L
 
 | | Crystal structure of the bromodomain of BRD9 in complex with compound 9. | | Descriptor: | 5-[3,5-dimethoxy-4-[(3-oxidanylazetidin-1-yl)methyl]phenyl]-1,3-dimethyl-pyridin-2-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
7RIP
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 soaked with CTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DAL
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, Nalpha-{(2S)-2-[(tert-butoxycarbonyl)amino]-3-phenylpropyl}-N-[2-(pyridin-3-yl)ethyl]-D-phenylalaninamide, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
8DLO
 
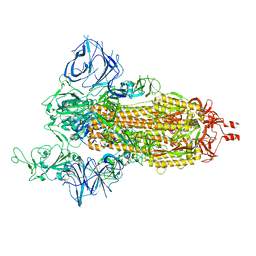 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLS
 
 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
9I97
 
 | | Cryo-EM structure of Shigella flexneri LptDE in complex with a Bicyclic Peptide binder (Compound 12) | | Descriptor: | 1-[4,7-bis(2-chloranylethanoyl)-1,4,7-triazonan-1-yl]-2-chloranyl-ethanone, Compound 12 - Bicyclic Peptide Binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, Hubbard, J, van den Berg, B, Newman, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
8DLT
 
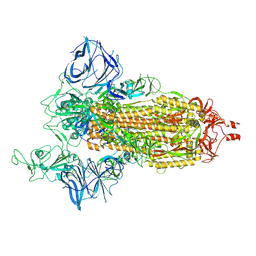 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
7RIY
 
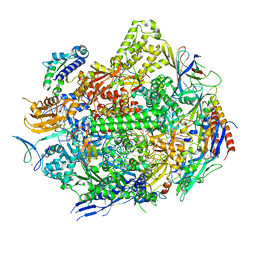 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 soaked with UTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6P1Q
 
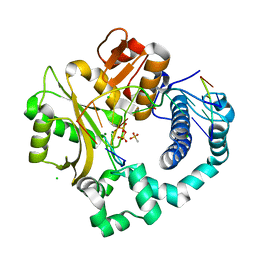 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated dCMP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*(8OG)P*TP*AP*CP*G)-3'), ... | | Authors: | Pedersen, L.C, Kaminski, A.M, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
5BJW
 
 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, T595S variant, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
9I98
 
 | | Cryo-EM structure of Shigella flexneri LptDE bound by a Bicyclic peptide molecule (Compound 13) | | Descriptor: | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone, Compound 13 - Bicyclic Peptide binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Newman, H, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, van den Berg, B, Hubbard, J. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
7ROQ
 
 | | Alternative Structure of Human ABCA1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aller, S.G. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Refined ABCA1 with Domain-swapped Regulatory Domains
Plos One, 2022
|
|
8U01
 
 | | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain Containing Protein from Phocaeicola plebeius | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain
Containing Protein from Phocaeicola plebeius
To Be Published
|
|
9I96
 
 | | Cryo-EM structure of Shigella flexneri LptDE bound by a Bicyclic peptide molecule (Compound 5) | | Descriptor: | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone, Compound 5 - Bicyclic Peptide binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Newman, H, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, van den Berg, B, Hubbard, J. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
