4DOI
 
 | |
4B9A
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BAL
 
 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxymethyltriazoledipicolinate complex at 1.30 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4KR1
 
 | |
4M2S
 
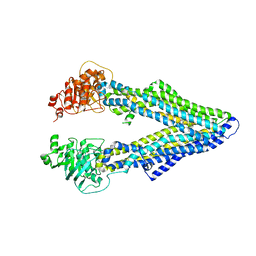 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
4DOK
 
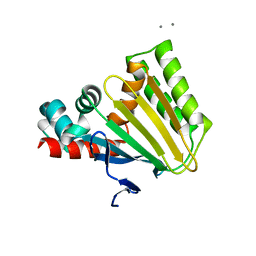 | |
4NX2
 
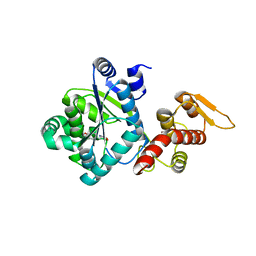 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4C5J
 
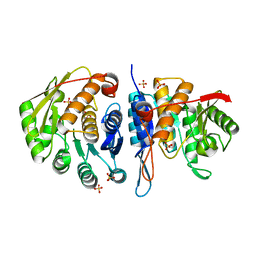 | | Structure of the pyridoxal kinase from Staphylococcus aureus | | Descriptor: | PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4EAM
 
 | | 1.70A resolution structure of apo beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
4EXA
 
 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4K83
 
 | | Crystal structure of lv-ranaspumin (Lv-RSN-1) from the foam nest of Leptodactylus vastus, orthorhombic crystal form | | Descriptor: | Lv-ranaspumin (Lv-RSN-1) | | Authors: | Hissa, D.C, Bezerra, G.A, Melo, V.M.M, Gruber, K. | | Deposit date: | 2013-04-17 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unique Crystal Structure of a Novel Surfactant Protein from the Foam Nest of the Frog Leptodactylus vastus.
Chembiochem, 15, 2014
|
|
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4ETR
 
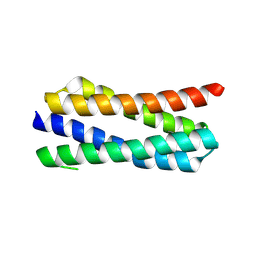 | | X-ray structure of PA2169 from Pseudomonas aeruginosa | | Descriptor: | Putative uncharacterized protein | | Authors: | Schnell, R, Sandalova, T, Lindqvist, Y, Schneider, G. | | Deposit date: | 2012-04-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4EXB
 
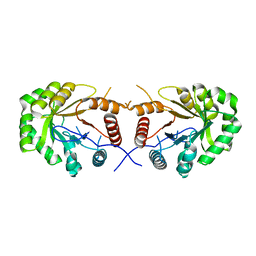 | |
4ES6
 
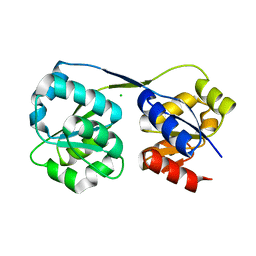 | |
4NXF
 
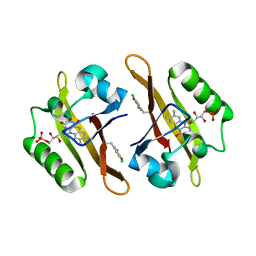 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXG
 
 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
6X8M
 
 | | CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
7PGB
 
 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PG8
 
 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RIV
 
 | | human PXR LBD bound to GSK001 | | Descriptor: | Dabrafenib, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ward, P. | | Deposit date: | 2021-07-20 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
