1E39
 
 | | Flavocytochrome C3 from Shewanella frigidimarina histidine 365 mutated to alanine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT, FUMARIC ACID, ... | | Authors: | Doherty, M.K, Pealing, S.L, Miles, C.S, Moysey, R, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2000-06-07 | | Release date: | 2000-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of the Active Site Acid/Base Catalyst in a Bacterial Fumarate Reductase: A Kinetic and Crystallographic Study
Biochemistry, 39, 2000
|
|
1E3A
 
 | | A slow processing precursor penicillin acylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hewitt, L, Kasche, V, Lummer, K, Lewis, R.J, Murshudov, G.N, Verma, C.S, Dodson, G.G, Wilson, K.S. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Slow Processing Precursor Penicillin Acylase from Escherichia Coli Reveals the Linker Peptide Blocking the Active-Site Cleft
J.Mol.Biol., 302, 2000
|
|
1E3B
 
 | | CYCLOPHILIN 3 FROM C.ELEGANS COMPLEXED WITH AUP(ET)3 | | Descriptor: | CYCLOPHILIN 3, GOLD ION, TRIETHYLPHOSPHANE | | Authors: | Zou, J, Taylor, P, Dornan, J, Robinson, S.P, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2000-06-08 | | Release date: | 2000-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First Crystal Structure of a Medicinally Relevant Gold Protein Complex:Unexpected Binding of [Au(Pet (3))](+) to Histidine
Angew.Chem.Int.Ed.Engl., 39, 2000
|
|
1E3C
 
 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-13 | | Release date: | 2001-03-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3D
 
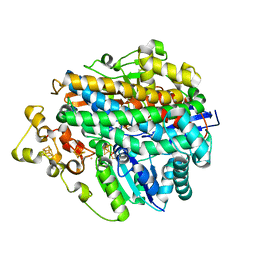 | | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), FE3-S4 CLUSTER, ... | | Authors: | Matias, P.M, Soares, C.M, Saraiva, L.M, Coelho, R, Morais, J, Le Gall, J, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3.
J.Biol.Inorg.Chem., 6, 2001
|
|
1E3E
 
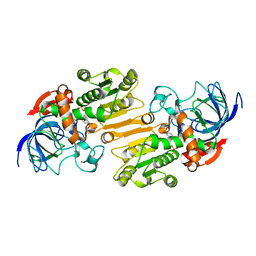 | | Mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoeoeg, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-14 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3F
 
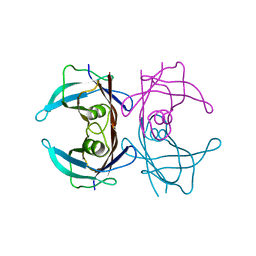 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-06-14 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E3G
 
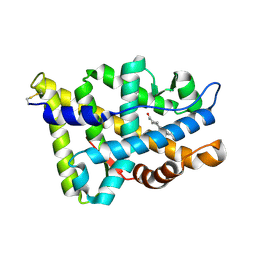 | | Human Androgen Receptor Ligand Binding in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, ANDROGEN RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Ruff, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3H
 
 | | SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | GUANOSINE PENTAPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-15 | | Release date: | 2000-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1E3I
 
 | | Mouse class II alcohol dehydrogenase complex with NADH and inhibitor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-16 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3J
 
 | | Ketose reductase (sorbitol dehydrogenase) from silverleaf whitefly | | Descriptor: | BORIC ACID, NADP(H)-DEPENDENT KETOSE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Banfield, M.J, Salvucci, M.E, Baker, E.N, Smith, C.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nadp(H)-Dependent Ketose Reductase from Besimia Argentifolii at 2.3 Angstrom Resolution
J.Mol.Biol., 306, 2001
|
|
1E3K
 
 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3L
 
 | | P47H mutant of mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | ALCOHOL DEHYDROGENASE, CLASS II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-19 | | Release date: | 2000-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3M
 
 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
1E3O
 
 | | Crystal structure of Oct-1 POU dimer bound to MORE | | Descriptor: | 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*A)-3', 5'-D(*TP*CP*CP*TP*CP*AP*TP*GP*CP*AP*T)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Schoeler, H, Wilmanns, M. | | Deposit date: | 2000-06-20 | | Release date: | 2001-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1E3P
 
 | | tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | Polyribonucleotide nucleotidyltransferase, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1E3Q
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE COMPLEXED WITH BW284C51 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Felder, C.E, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a Complex of the Potent and Specific Inhibitor Bw284C51 with Torpedo Californica Acetylcholinesterase
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1E3R
 
 | | Crystal structure of ketosteroid isomerase mutant D40N (D38N TI numbering) from Pseudomonas putida complexed with androsten-3beta-ol-17-one | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Hyun, B.-H, Oh, B.-H. | | Deposit date: | 2000-06-22 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1E3S
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Powell, A.J, Read, J.A, Banfield, M.J, Brady, R.L. | | Deposit date: | 2000-06-22 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
1E3T
 
 | | Solution Structure of the NADP(H) binding Component (dIII) of Proton-Translocating Transhydrogenase from Rhodospirillum rubrum | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE (SUBUNIT BETA) | | Authors: | Jeeves, M, Smith, K.J, Quirk, P.G, Cotton, N.P.J, Jackson, J.B. | | Deposit date: | 2000-06-22 | | Release date: | 2000-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nadp(H)-Binding Component (Diii) of Proton-Translocating Transhydrogenase from Rhodospirillum Rubrum
Biochim.Biophys.Acta, 1459, 2000
|
|
1E3U
 
 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1E3V
 
 | | Crystal structure of ketosteroid isomerase from Psedomonas putida complexed with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Kim, J.-S, Oh, B.-H. | | Deposit date: | 2000-06-24 | | Release date: | 2001-03-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1E3W
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH and 3-keto butyrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Powell, A.J, Read, J.A, Brady, R.L. | | Deposit date: | 2000-06-26 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
1E3X
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E3Y
 
 | |
