6VT8
 
 | | Naegleria gruberi RNA ligase E312A mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
7A33
 
 | |
1KGY
 
 | | Crystal Structure of the EphB2-ephrinB2 complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2, EPHRIN-B2 | | Authors: | Himanen, J.P, Rajashankar, K.R, Lackmann, M, Cowan, C.A, Henkemeyer, M, Nikolov, D.B. | | Deposit date: | 2001-11-28 | | Release date: | 2002-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an Eph receptor-ephrin complex.
Nature, 414, 2001
|
|
1KEL
 
 | |
7A2W
 
 | |
7A3A
 
 | |
1KEX
 
 | | Crystal Structure of the b1 Domain of Human Neuropilin-1 | | Descriptor: | Neuropilin-1 | | Authors: | Lee, C.C, Kreusch, A, McMullan, D, Ng, K, Spraggon, G. | | Deposit date: | 2001-11-18 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Neuropilin-1 b1 Domain
Structure, 11, 2003
|
|
1KHD
 
 | | Crystal Structure Analysis of the anthranilate phosphoribosyltransferase from Erwinia carotovora at 1.9 resolution (current name, Pectobacterium carotovorum) | | Descriptor: | Anthranilate phosphoribosyltransferase | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
1KI3
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH PENCICLOVIR | | Descriptor: | 9-(4-HYDROXY-3-(HYDROXYMETHYL)BUT-1-YL)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Jarvest, R.L, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KIS
 
 | |
7ACO
 
 | |
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KFE
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH L-Ser Bound To The Beta Site | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
6PD4
 
 | | Crystal Structure of Hendra Virus Attachment G Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New insights into the Hendra virus attachment and entry process from structures of the virus G glycoprotein and its complex with Ephrin-B2.
PLoS ONE, 7, 2012
|
|
7ACL
 
 | |
7ACM
 
 | |
1KIY
 
 | | D100E trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, trichodiene synthase | | Authors: | Rynkiewicz, M.J, Cane, D.E, Christianson, D.W. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity.
Biochemistry, 41, 2002
|
|
2XD1
 
 | | ACTIVE SITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7A0S
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
1KJK
 
 | |
1KK9
 
 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
1KKJ
 
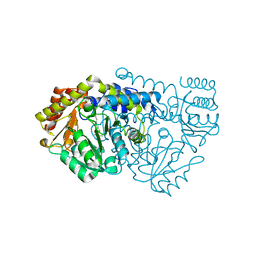 | | Crystal Structure of Serine Hydroxymethyltransferase from B.stearothermophilus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
7A2P
 
 | |
1KFP
 
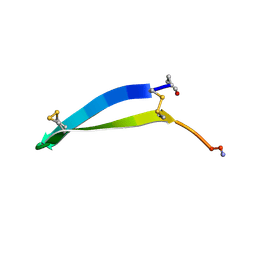 | | Solution structure of the antimicrobial 18-residue gomesin | | Descriptor: | GOMESIN | | Authors: | Mandard, N, Bulet, P, Caille, A, Daffre, S, Vovelle, F. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider.
Eur.J.Biochem., 269, 2002
|
|
1KFY
 
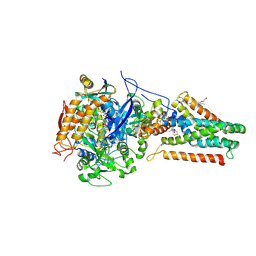 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
