1HFZ
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Pike, A.C.W, Brew, K, Acharya, K.R. | | Deposit date: | 1996-06-13 | | Release date: | 1997-07-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase.
Structure, 4, 1996
|
|
1JSU
 
 | | P27(KIP1)/CYCLIN A/CDK2 COMPLEX | | Descriptor: | CYCLIN A, CYCLIN-DEPENDENT KINASE-2, P27, ... | | Authors: | Russo, A.A, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1996-07-03 | | Release date: | 1997-07-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex.
Nature, 382, 1996
|
|
1LIA
 
 | |
313D
 
 | | Z-DNA HEXAMER WITH 5' OVERHANGS THAT FORM A REVERSE HOOGSTEEN BASE PAIR | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(5CM)P*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Mooers, B.H.M, Eichman, B.F, Ho, P.S. | | Deposit date: | 1997-02-04 | | Release date: | 1997-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structures and relative stabilities of d(G x G) reverse Hoogsteen, d(G x T) reverse wobble, and d(G x C) reverse Watson-Crick base-pairs in DNA crystals.
J.Mol.Biol., 269, 1997
|
|
314D
 
 | | Z-DNA HEXAMER WITH 5' OVERHANGS THAT FORM A REVERSE WOBBLE BASE PAIR | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Mooers, B.H.M, Eichman, B.F, Ho, P.S. | | Deposit date: | 1997-02-04 | | Release date: | 1997-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures and relative stabilities of d(G x G) reverse Hoogsteen, d(G x T) reverse wobble, and d(G x C) reverse Watson-Crick base-pairs in DNA crystals.
J.Mol.Biol., 269, 1997
|
|
1MVI
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA, NMR, 15 STRUCTURES | | Descriptor: | MVIIA | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1VII
 
 | |
1MVJ
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1QIL
 
 | |
1KBC
 
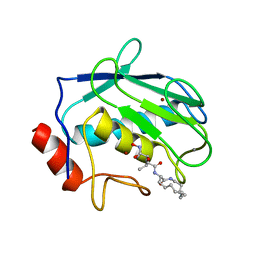 | | PROCARBOXYPEPTIDASE TERNARY COMPLEX | | Descriptor: | 3-AMINO-AZACYCLOTRIDECAN-2-ONE, 3-FORMYL-2-HYDROXY-5-METHYL-HEXANOIC ACID HYDROXYAMIDE, CALCIUM ION, ... | | Authors: | Betz, M, Gomis-Rueth, F.X, Bode, W. | | Deposit date: | 1997-04-29 | | Release date: | 1997-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8-A crystal structure of the catalytic domain of human neutrophil collagenase (matrix metalloproteinase-8) complexed with a peptidomimetic hydroxamate primed-side inhibitor with a distinct selectivity profile.
Eur.J.Biochem., 247, 1997
|
|
1CJL
 
 | |
1AB1
 
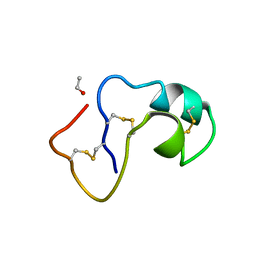 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
1BTV
 
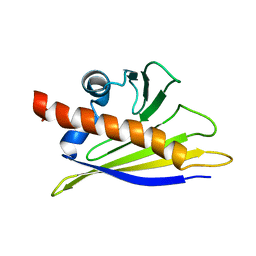 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1TSK
 
 | |
2QIL
 
 | |
3GRT
 
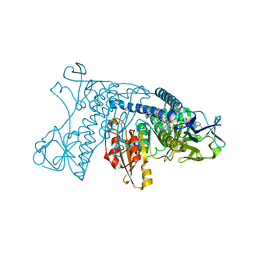 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED TRYPANOTHIONE COMPLEX | | Descriptor: | 2-AMINO-4-[4-(4-AMINO-4-CARBOXY-BUTYRYLAMINO)-5,8,19,22-TETRAOXO-1,2-DITHIA-6,9,13,18,21-PENTAAZA-CYCLOTETRACOS-23-YLCARBAMOYL]-BUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1DPM
 
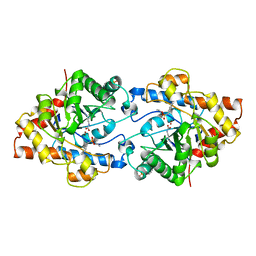 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIETHYL 4-METHYLBENZYLPHOSPHONATE | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, FORMIC ACID, PHOSPHOTRIESTERASE, ... | | Authors: | Vanhooke, J.L, Benning, M.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 1996-02-13 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
Biochemistry, 35, 1996
|
|
1AFZ
 
 | |
1AFF
 
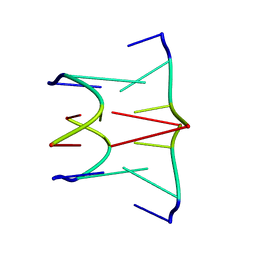 | | DNA QUADRUPLEX CONTAINING GGGG TETRADS AND (T.A).A TRIADS, NMR, 8 STRUCTURES | | Descriptor: | QUADRUPLEX DNA (5'-D(TP*AP*GP*G)-3') | | Authors: | Kettani, A, Bouaziz, S, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads.
Nat.Struct.Biol., 4, 1997
|
|
1EMC
 
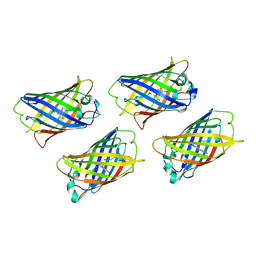 | |
1EMK
 
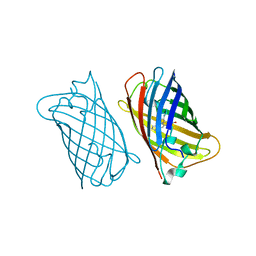 | |
1EMF
 
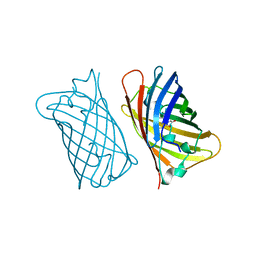 | |
1EME
 
 | |
2TDM
 
 | | STRUCTURE OF THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1997-03-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
1FDI
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI COMPLEXED WITH THE INHIBITOR NITRITE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
