5JIO
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase ternary complex with ADP and Glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
6ZB3
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, PENTAETHYLENE GLYCOL, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
4U7J
 
 | |
5JD9
 
 | | Bacillus cereus CotH kinase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Spore coat protein H | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Phosphorylation of spore coat proteins by a family of atypical protein kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | Descriptor: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9BF8
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) Untagged Crystal Structure | | Descriptor: | 1,2-ETHANEDIOL, ORF1a polyprotein, PHOSPHATE ION, ... | | Authors: | Al-Homoudi, A.I, Engel, J, Brunzelle, J.S, Gavande, N, Kovari, L.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human Structural Homologues of SARS-CoV-2 PL pro as Anti-Targets: A Strategic Panel Analysis.
MicroPubl Biol, 2025, 2025
|
|
9GN8
 
 | | Crystal Structure of UFC1 E149D | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Kumar, M, Banerjee, S, Isupov, M.N, Wiener, R. | | Deposit date: | 2024-08-31 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
4TZ8
 
 | | Structure of human ATAD2 bromodomain bound to fragment inhibitor | | Descriptor: | 2-amino-7,7-dimethyl-5,6,7,8-tetrahydro-4H-[1,3]thiazolo[5,4-c]azepin-4-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
6ZMA
 
 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 140 bar of krypton using the soak and freeze methodology | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
9C0P
 
 | | M. tuberculosis PKS13 acyltransferase (AT) domain in complex with SuFEx inhibitor CEC215 | | Descriptor: | N-{4-[(4-{[fluorodi(hydroxy)-lambda~4~-sulfanyl]oxy}phenoxy)methyl]phenyl}acetamide, PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, ... | | Authors: | Krieger, I.V, Tang, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-05-27 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13.
Nature, 645, 2025
|
|
8C3W
 
 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6YVS
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | Descriptor: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
8XVR
 
 | | Crystal structure of inulosucrase from Lactobacillus reuteri 121 mutant R544W | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ni, D, Hou, X, Cheng, M, Xu, W, Rao, Y, Mu, W. | | Deposit date: | 2024-01-15 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided tunnel engineering to reveal the molecular basis of sugar chain extension of inulosucrase
To Be Published
|
|
4U0W
 
 | |
6FYK
 
 | | X-Ray structure of CLK2-KD(136-496)/Indazole1 at 2.39A | | Descriptor: | (2~{S})-4-methyl-1-[5-(3-methyl-2~{H}-indazol-5-yl)pyridin-3-yl]oxy-pentan-2-amine, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
5FEH
 
 | | Crystal structure of PCT64_35B, a broadly neutralizing anti-HIV antibody | | Descriptor: | 1,2-ETHANEDIOL, PCT64_26 Fab heavy chain, PCT64_26 Fab light chain, ... | | Authors: | Murrell, S, Wilson, I.A. | | Deposit date: | 2015-12-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage.
Immunity, 47, 2017
|
|
8F9S
 
 | |
6FQ1
 
 | |
8F1Z
 
 | | EGFR kinase in complex with Bayer #33 | | Descriptor: | (2P)-3-(3-chloro-2-methoxyanilino)-2-[3-(2-methoxy-2-methylpropoxy)pyridin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5L9O
 
 | |
9GRG
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease in complex with PMSF | | Descriptor: | Alkaline serine protease, CALCIUM ION, GLYCEROL, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, Windhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2024-09-11 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
8XSD
 
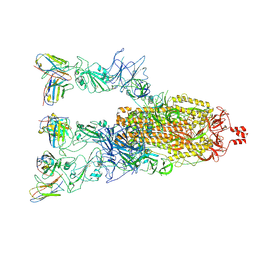 | | BA.5 Spike complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike glycoprotein | | Authors: | Feng, L.L. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
9F96
 
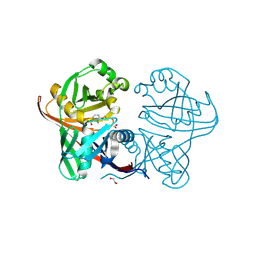 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-ethoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-3-ethoxy-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 19, 2024
|
|
8F1X
 
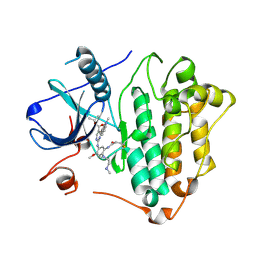 | | EGFR kinase in complex with mobocertinib (TAK-788) | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7XC4
 
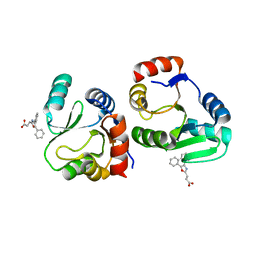 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
