1DTK
 
 | |
1DTL
 
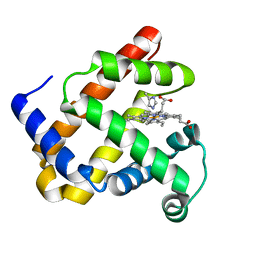
 | | CRYSTAL STRUCTURE OF CALCIUM-SATURATED (3CA2+) CARDIAC TROPONIN C COMPLEXED WITH THE CALCIUM SENSITIZER BEPRIDIL AT 2.15 A RESOLUTION | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, CARDIAC TROPONIN C | | Authors: | Li, Y, Love, M.L, Putkey, J.A, Cohen, C. | | Deposit date: | 2000-01-12 | | Release date: | 2000-05-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Bepridil opens the regulatory N-terminal lobe of cardiac troponin C.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DTM
 
 | |
1DTN
 
 | |
1DTO
 
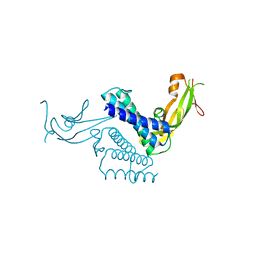 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1DTP
 
 | |
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTS
 
 | | CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE, DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-03-28 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 A resolution.
Structure, 2, 1994
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTU
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE: A MUTANT Y89D/S146P COMPLEXED TO AN HEXASACCHARIDE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production.
J.Mol.Biol., 296, 2000
|
|
1DTV
 
 | | NMR STRUCTURE OF THE LEECH CARBOXYPEPTIDASE INHIBITOR (LCI) | | Descriptor: | CARBOXYPEPTIDASE INHIBITOR | | Authors: | Reverter, D, Fernandez-Catalan, C, Bode, W, Holak, T.A, Aviles, F.X. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2.
Nat.Struct.Biol., 7, 2000
|
|
1DTW
 
 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | Authors: | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
1DTX
 
 | |
1DTY
 
 | | CRYSTAL STRUCTURE OF ADENOSYLMETHIONINE-8-AMINO-7-OXONANOATE AMINOTRANSFERASE WITH PYRIDOXAL PHOSPHATE COFACTOR. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Alexeev, D, Sawyer, L, Baxter, R.L, Alexeeva, M.V, Campopiano, D. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of adenosylmethionine-8-amino-7-oxonanoate aminotransferase with pyridoxal phosphate cofactor
to be published
|
|
1DTZ
 
 | | STRUCTURE OF CAMEL APO-LACTOFERRIN DEMONSTRATES ITS DUAL ROLE IN SEQUESTERING AND TRANSPORTING FERRIC IONS SIMULTANEOUSLY:CRYSTAL STRUCTURE OF CAMEL APO-LACTOFERRIN AT 2.6A RESOLUTION. | | Descriptor: | APO LACTOFERRIN | | Authors: | Khan, J.A, Kumar, P, Paramasivam, M, Srinivasan, A, Yadav, R.S, Sahani, M.S, Singh, T.P. | | Deposit date: | 2000-01-13 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Camel lactoferrin, a transferrin-cum-lactoferrin: crystal structure of camel apolactoferrin at 2.6 A resolution and structural basis of its dual role.
J.Mol.Biol., 309, 2001
|
|
1DU0
 
 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1DU1
 
 | |
1DU2
 
 | | SOLUTION STRUCTURE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Descriptor: | DNA POLYMERASE III | | Authors: | Keniry, M.A, Berthon, H.A, Yang, J.-Y, Miles, C.S, Dixon, N.E. | | Deposit date: | 2000-01-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli.
Protein Sci., 9, 2000
|
|
1DU3
 
 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
1DU4
 
 | |
1DU5
 
 | | THE CRYSTAL STRUCTURE OF ZEAMATIN. | | Descriptor: | ZEAMATIN | | Authors: | Batalia, M.A, Monzingo, A.F, Ernst, S, Roberts, W, Robertus, J.D. | | Deposit date: | 2000-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family.
Nat.Struct.Biol., 3, 1996
|
|
1DU6
 
 | |
1DU9
 
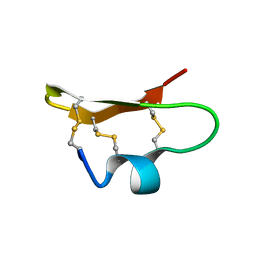 | | SOLUTION STRUCTURE OF BMP02, A NATURAL SCORPION TOXIN WHICH BLOCKS APAMIN-SENSITIVE CALCIUM-ACTIVATED POTASSIUM CHANNELS, 25 STRUCTURES | | Descriptor: | BMP02 NEUROTOXIN | | Authors: | Xu, Y, Wu, J, Pei, J, Shi, Y, Ji, Y, Tong, Q. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP02, a new potassium channel blocker from the venom of the Chinese scorpion Buthus martensi Karsch.
Biochemistry, 39, 2000
|
|
1DUA
 
 | |
1DUB
 
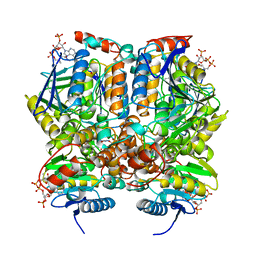 | | 2-ENOYL-COA HYDRATASE, DATA COLLECTED AT 100 K, PH 6.5 | | Descriptor: | 2-ENOYL-COA HYDRATASE, ACETOACETYL-COENZYME A | | Authors: | Wierenga, R.K, Engel, C.K. | | Deposit date: | 1996-06-10 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket.
EMBO J., 15, 1996
|
|
